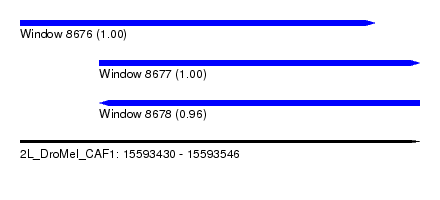
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,593,430 – 15,593,546 |
| Length | 116 |
| Max. P | 0.999878 |

| Location | 15,593,430 – 15,593,533 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15593430 103 + 22407834 CCGAACAUCUGUAUUAAUUUACGGAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCA .(((...((((((......))))))(((((..(((.(((((((.(((((....))))).)))))))..)))))))))))(((.(((((....)))))..))). ( -36.30) >DroSec_CAF1 100054 103 + 1 CCGAACAUCUGUAUUAAUUUACGGAUUUGGAUUUAAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCA .(((...((((((......))))))(((((.(((..(((((((.(((((....))))).)))))))..)))))))))))(((.(((((....)))))..))). ( -36.70) >DroSim_CAF1 92147 103 + 1 CCGAACAUCUGUAUUAAUUUACGGAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCA .(((...((((((......))))))(((((..(((.(((((((.(((((....))))).)))))))..)))))))))))(((.(((((....)))))..))). ( -36.30) >DroEre_CAF1 94223 103 + 1 CCCAAUUUCUGUAUUAAUUUACGGAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGAUCAAAACGGACAGCUGACAGGUCAACCGGUCG (((((..((((((......)))))).)))(((((..(((((((.(((((....))))).)))))))..))))).....))......(((.((....)).))). ( -29.80) >DroYak_CAF1 97850 103 + 1 CCCAACAUCUGUAUUAAUUUACGGACUUGGAUUUCAAUUGCUUAUUGGCCUGGGUCAGAGAGCAAUGCGGACCAAAUCAGACAGCUGACAGGUCAACCGGUUG ..((((.((((((......))))))..(((......(((((((.(((((....))))).)))))))...((((...((((....))))..))))..))))))) ( -32.80) >consensus CCGAACAUCUGUAUUAAUUUACGGAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCA .......((((((......))))))(((((.(((..(((((((.(((((....))))).)))))))..))))))))...(((.(((((....)))))..))). (-29.90 = -30.18 + 0.28)

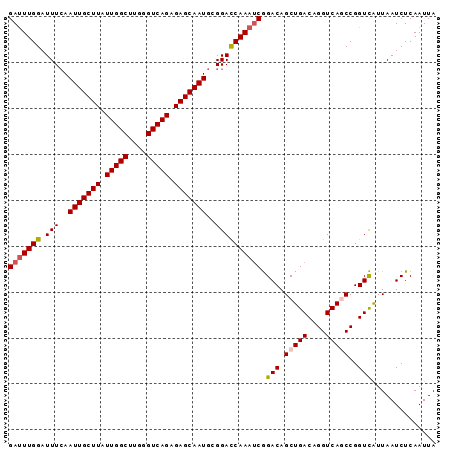
| Location | 15,593,453 – 15,593,546 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15593453 93 + 22407834 GAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCAUUAAUCUCAAUUA (((((((..(((.(((((((.(((((....))))).)))))))..)))))))))).(((.(((((....)))))..))).............. ( -33.20) >DroSec_CAF1 100077 93 + 1 GAUUUGGAUUUAAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCAUUAAUCUCAAUUA (((((((.(((..(((((((.(((((....))))).)))))))..)))))))))).(((.(((((....)))))..))).............. ( -33.60) >DroSim_CAF1 92170 93 + 1 GAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCAUUAAUCUCAAUUA (((((((..(((.(((((((.(((((....))))).)))))))..)))))))))).(((.(((((....)))))..))).............. ( -33.20) >DroEre_CAF1 94246 93 + 1 GAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGAUCAAAACGGACAGCUGACAGGUCAACCGGUCGUUAAUCUCAAUUA ((((..(((((..(((((((.(((((....))))).)))))))..)))))..((((..(.(.(((....))).).)..))))))))....... ( -23.60) >DroYak_CAF1 97873 93 + 1 GACUUGGAUUUCAAUUGCUUAUUGGCCUGGGUCAGAGAGCAAUGCGGACCAAAUCAGACAGCUGACAGGUCAACCGGUUGUUAAUCUCAAUUA ((.((((..(((.(((((((.(((((....))))).)))))))..))))))).)).((((((((..........))))))))........... ( -27.70) >consensus GAUUUGGAUUUCAAUUGCUUAUUGGCUUGGGUCAGAGAGCAAUGCGGACCAAAUCGGACAGCUGACAGGUCAGCCGGUCAUUAAUCUCAAUUA (((((((.(((..(((((((.(((((....))))).)))))))..)))))))))).(((.(((((....)))))..))).............. (-28.26 = -28.74 + 0.48)

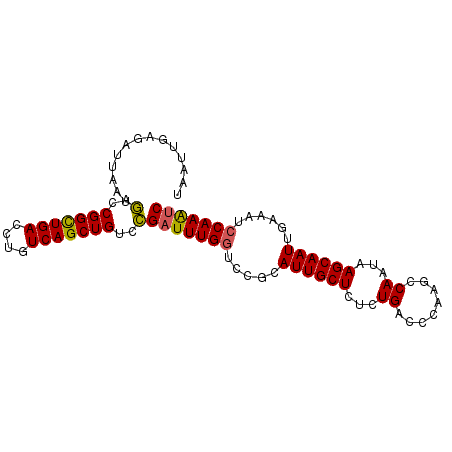
| Location | 15,593,453 – 15,593,546 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
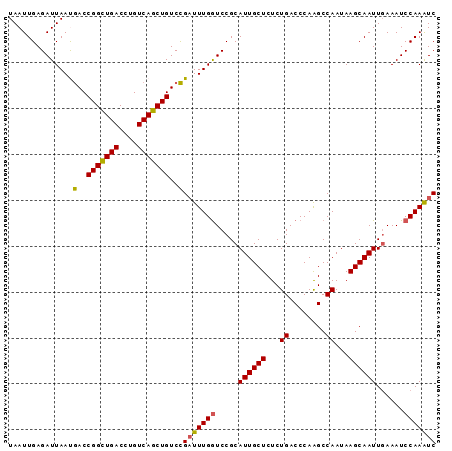
>2L_DroMel_CAF1 15593453 93 - 22407834 UAAUUGAGAUUAAUGACCGGCUGACCUGUCAGCUGUCCGAUUUGGUCCGCAUUGCUCUCUGACCCAAGCCAAUAAGCAAUUGAAAUCCAAAUC ..............(..(((((((....)))))))..)(((((((....(((((((...((........))...))))).))....))))))) ( -24.10) >DroSec_CAF1 100077 93 - 1 UAAUUGAGAUUAAUGACCGGCUGACCUGUCAGCUGUCCGAUUUGGUCCGCAUUGCUCUCUGACCCAAGCCAAUAAGCAAUUUAAAUCCAAAUC ..............(..(((((((....)))))))..)(((((((.....((((((...((........))...))))))......))))))) ( -23.20) >DroSim_CAF1 92170 93 - 1 UAAUUGAGAUUAAUGACCGGCUGACCUGUCAGCUGUCCGAUUUGGUCCGCAUUGCUCUCUGACCCAAGCCAAUAAGCAAUUGAAAUCCAAAUC ..............(..(((((((....)))))))..)(((((((....(((((((...((........))...))))).))....))))))) ( -24.10) >DroEre_CAF1 94246 93 - 1 UAAUUGAGAUUAACGACCGGUUGACCUGUCAGCUGUCCGUUUUGAUCCGCAUUGCUCUCUGACCCAAGCCAAUAAGCAAUUGAAAUCCAAAUC ...(((.((((((((..(((((((....)))))))..))))......((.((((((...((........))...)))))))).)))))))... ( -20.20) >DroYak_CAF1 97873 93 - 1 UAAUUGAGAUUAACAACCGGUUGACCUGUCAGCUGUCUGAUUUGGUCCGCAUUGCUCUCUGACCCAGGCCAAUAAGCAAUUGAAAUCCAAGUC .................(((((((....)))))))...(((((((....(((((((...((.(....).))...))))).))....))))))) ( -21.60) >consensus UAAUUGAGAUUAAUGACCGGCUGACCUGUCAGCUGUCCGAUUUGGUCCGCAUUGCUCUCUGACCCAAGCCAAUAAGCAAUUGAAAUCCAAAUC ..............(..(((((((....)))))))..)(((((((.....((((((...((........))...))))))......))))))) (-20.64 = -20.32 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:53 2006