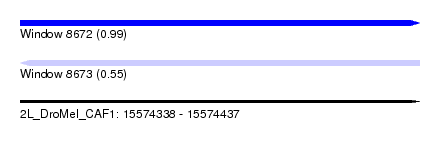
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,574,338 – 15,574,437 |
| Length | 99 |
| Max. P | 0.986206 |

| Location | 15,574,338 – 15,574,437 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
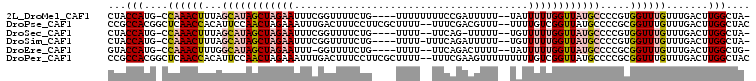
>2L_DroMel_CAF1 15574338 99 + 22407834 -UAGCCAAGUCAAACAAACCACGGGGCAUAACCAAAAAUA--AAAAAUCGGAAAAAAAA----CAGAAAACCGAAAUUCUAGCUAUGCUAAAGUUUGG-CAUGGUAG -..((((.(((((((........((......)).......--.....((((........----.......)))).....((((...))))..))))))-).)))).. ( -20.66) >DroPse_CAF1 135447 103 + 1 GUAGCCAAGUCAAACAAACCGCGGGGCAUAACCGACAAAA--AAACGUCGAAA--AAAAGCGAAGGAAAGUCAAAUUUCUAGUUGGAAUGUGGUUGAGCCGUGGCGG ..................((((.((((.(((((.(((...--.(((.(((...--.....))).(((((......))))).)))....)))))))).))).).)))) ( -25.80) >DroSec_CAF1 87959 96 + 1 -UAGCCAAGUCAAACAAACCACGGGGCAUAACCAAAAACA--AAAAA-CUGAA--AAAA----CAGAAAACCGAAAUUCUAGCUAUGCUAAAGUUUGG-CAUGGUAG -..((((.(((((((......(((((.....)).......--.....-(((..--....----)))....)))......((((...))))..))))))-).)))).. ( -22.70) >DroSim_CAF1 79898 98 + 1 -UAGCCAAGUCAAACAAACCACGGGGCAUAACCAAAAACA--AAAAAUCUGAAA-AAAA----CAGAAAACCGAAAUUCUAGCUAUGCUAAAGUUUGG-CAUGGUAG -..((((.(((((((......(((((.....)).......--.....((((...-....----))))...)))......((((...))))..))))))-).)))).. ( -23.60) >DroEre_CAF1 81889 96 + 1 -CAGCCAAGUCAAACAAACCGCGGGGCAUAACCAAAAAUA--AAAAGUCUGAA--AAAA----CAGAAAACC-AAAUUCUAGCUAUGCCAAAGUUUGG-CAUGGUAC -..((((.(((((((...(....)((((((..........--.....((((..--....----)))).....-..........))))))...))))))-).)))).. ( -25.01) >DroPer_CAF1 134621 105 + 1 GUAGCCAAGUCAAACAAACCGCGGGGCAUAACCGACAAAAAAAAACUUCGAAA--AAAAGCGAAGGAAAGUCAAAUUUCUAGUUGGAAUGUGGUUGAGCCGUGGCGG ..................((((.((((.(((((.(((........(((((...--.....)))))((((......)))).........)))))))).))).).)))) ( -26.90) >consensus _UAGCCAAGUCAAACAAACCACGGGGCAUAACCAAAAAAA__AAAAAUCGGAA__AAAA____CAGAAAACCGAAAUUCUAGCUAUGCUAAAGUUUGG_CAUGGUAG ...((((.(((((((........((......))...............................((((........))))............)))))).).)))).. (-12.48 = -12.70 + 0.22)


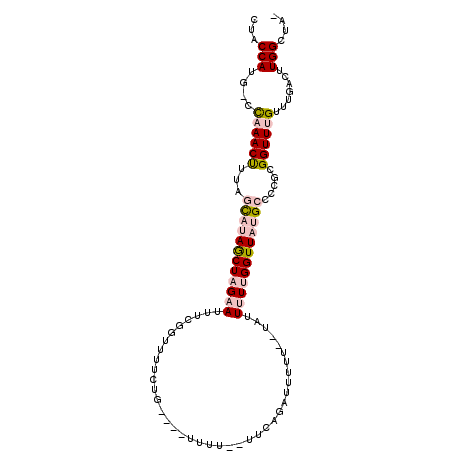
| Location | 15,574,338 – 15,574,437 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -10.27 |
| Energy contribution | -11.38 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15574338 99 - 22407834 CUACCAUG-CCAAACUUUAGCAUAGCUAGAAUUUCGGUUUUCUG----UUUUUUUUCCGAUUUUU--UAUUUUUGGUUAUGCCCCGUGGUUUGUUUGACUUGGCUA- .......(-((((......((((((((((((..((((.......----........)))).....--...))))))))))))......(((.....))))))))..- ( -21.66) >DroPse_CAF1 135447 103 - 1 CCGCCACGGCUCAACCACAUUCCAACUAGAAAUUUGACUUUCCUUCGCUUUU--UUUCGACGUUU--UUUUGUCGGUUAUGCCCCGCGGUUUGUUUGACUUGGCUAC ..((((.((.((((..(((..((.....((((......))))....((...(--..((((((...--...))))))..).)).....))..)))))))))))))... ( -21.60) >DroSec_CAF1 87959 96 - 1 CUACCAUG-CCAAACUUUAGCAUAGCUAGAAUUUCGGUUUUCUG----UUUU--UUCAG-UUUUU--UGUUUUUGGUUAUGCCCCGUGGUUUGUUUGACUUGGCUA- .......(-((((......((((((((((((...(((....(((----....--..)))-....)--)).))))))))))))......(((.....))))))))..- ( -23.10) >DroSim_CAF1 79898 98 - 1 CUACCAUG-CCAAACUUUAGCAUAGCUAGAAUUUCGGUUUUCUG----UUUU-UUUCAGAUUUUU--UGUUUUUGGUUAUGCCCCGUGGUUUGUUUGACUUGGCUA- .......(-((((......((((((((((((...(((...((((----....-...))))....)--)).))))))))))))......(((.....))))))))..- ( -23.90) >DroEre_CAF1 81889 96 - 1 GUACCAUG-CCAAACUUUGGCAUAGCUAGAAUUU-GGUUUUCUG----UUUU--UUCAGACUUUU--UAUUUUUGGUUAUGCCCCGCGGUUUGUUUGACUUGGCUG- .......(-((((.....(((((((((((((...-.....((((----....--..)))).....--...))))))))))))).....(((.....))))))))..- ( -26.59) >DroPer_CAF1 134621 105 - 1 CCGCCACGGCUCAACCACAUUCCAACUAGAAAUUUGACUUUCCUUCGCUUUU--UUUCGAAGUUUUUUUUUGUCGGUUAUGCCCCGCGGUUUGUUUGACUUGGCUAC ..((((.(((..(((((((.........((((......))))(((((.....--...)))))........))).))))..)))....((((.....))))))))... ( -21.60) >consensus CUACCAUG_CCAAACUUUAGCAUAGCUAGAAUUUCGGUUUUCUG____UUUU__UUCAGAUUUUU__UAUUUUUGGUUAUGCCCCGCGGUUUGUUUGACUUGGCUA_ ...(((....((((((...((((((((((((.......................................)))))))))))).....)))))).......))).... (-10.27 = -11.38 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:48 2006