
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,568,395 – 15,568,494 |
| Length | 99 |
| Max. P | 0.729990 |

| Location | 15,568,395 – 15,568,494 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -8.49 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
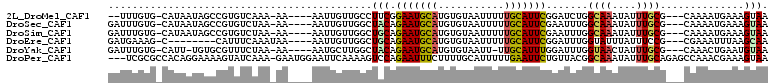
>2L_DroMel_CAF1 15568395 99 + 22407834 --UUUGUG-CAUAAUAGCCGUGUCAAA-AA----AAUUGUUGCCUUCGGAAUGCAUGUGUAAUUUUUGCAUUCGGAUCUGGCAAAUAUUUGCG---CAAAAUGAAAGUAA --((((((-((.((((.......(((.-..----..)))(((((.((.(((((((...........))))))).))...))))).))))))))---)))).......... ( -26.90) >DroSec_CAF1 82163 101 + 1 GAUUUGUG-CAUAAUAGCCGUGUCUAA-AA----AAUUGUUGGCUACAGAAUGCAUGUGUAAUUUUUGCAUUCGAAUUUGGCAAAUAUUUGCG---CAAAAUGAAAGUAA ..((((((-((.((((((((((.((((-..----.....)))).))).(((((((...........)))))))......)))...))))))))---)))).......... ( -25.30) >DroSim_CAF1 74117 101 + 1 GAUUUGUG-CAUAAUAGCCGUGUCUAA-AA----AAUUGUUGGCUGCAGAAUGCAUGUGUAAUUUUUGCAUUCGAAUUUGGCAAAUAUUUGCG---CAAAAUGAAAGUAA ..((((((-((.((((((((..((...-..----...(((.....)))(((((((...........)))))))))...))))...))))))))---)))).......... ( -24.80) >DroEre_CAF1 75982 94 + 1 GAUGAAAG-C--------CAUUUCAAAUAA----AAUUGUUGGCUGCAGAAUGCAUGUGUAAUUUUUGCAUUCGGAUUUGGUAUUUAUUUCCG---CGAAAUUUAAGCAA ......((-(--------((...(((....----..))).)))))((.(((((((...........)))))))(((..((.....))..))))---)............. ( -20.70) >DroYak_CAF1 79486 99 + 1 GAUUUGUG-CAUU-UGUGCGUUUCUAA-AA----AAUGCUUGGCUACAGAAUGCAUGUGUAAUU-UUGCAUUUGGAUUUGGUAACUAUUUGCG---CAAACUGAAUGUAA .....(((-((((-(..((((((....-.)----))))).((....))))))))))........-(((((((..(.((((((((....)))).---)))))..))))))) ( -24.30) >DroPer_CAF1 130086 106 + 1 ---UCGCGCCACAGGAAAAGUAUCAAA-GAAUGGAAUUCAAAAGUCCAGAAUUUCUUUUGCAUUUUUGAAUUCUGUUACGGCAAAUAUUUGCAGAGCCAAACGAAAGUAA ---....(((....((......))...-.((((((((((((((((.(((((....))))).))))))))))))))))..)))..................((....)).. ( -25.10) >consensus GAUUUGUG_CAUAAUAGCCGUGUCAAA_AA____AAUUGUUGGCUACAGAAUGCAUGUGUAAUUUUUGCAUUCGGAUUUGGCAAAUAUUUGCG___CAAAAUGAAAGUAA ............................................(((.(((((((...........))))))).......((((....))))..............))). ( -8.49 = -8.47 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:45 2006