| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,546,691 – 15,546,799 |
| Length | 108 |
| Max. P | 0.642944 |

| Location | 15,546,691 – 15,546,799 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
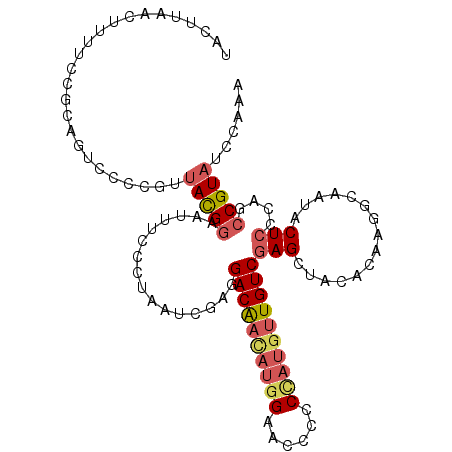
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
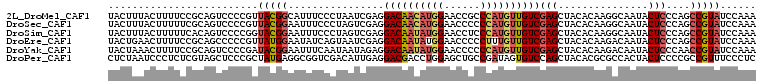
>2L_DroMel_CAF1 15546691 108 + 22407834 UACUUUACUUUUCCGCAGUCCCCGUUACGGCAUUUCCCUAAUCGAGGACAACAUGGAACCGCCCAUGUUGUCGAGCUACACAAGGCAAUACUCCCAGCCGUAUCCAAA .........................((((((...............((((((((((......))))))))))..(((......)))..........))))))...... ( -28.90) >DroSec_CAF1 53671 108 + 1 UACUUUACUUUUUCGCAGUCCCCGUUACGGAAUUUCCCUAGUCGAGGACAACAUGGAACCCCCCAUGUUGUCGAGCUACACAAGGCAAUACUCCCAGCCGUAUCCAAA .....(((...(((((.((((.(((((.((.....)).))).)).))))(((((((......)))))))).))))........(((..........))))))...... ( -26.10) >DroSim_CAF1 52337 108 + 1 UACUUUACUUUUUCACAGUCCCCGGUACGGAAUUUCCCUAGUCGAGGACAAUAUGGAACCUCCCAUGUUGUCGAGCUACACAAGGCAAUACUCCCAGCCGUAUCCAAA .......................(((((((.............(((((((((((((......))))))))))..(((......)))....)))....))))))).... ( -26.73) >DroEre_CAF1 54630 108 + 1 UACUGAACUUUUCCGCAGCCCCCGUUAUGGAAUAUCAGUAAUCGAGGACAAUAUGGAACCCCCUUUGUUGUCGAGCUACACAAGACAAUACUCCCAGCCGUAUCCAAA ((((((....(((((.(((....))).)))))..)))))).....(((...(((((.........(((((((...........))))))).......))))))))... ( -24.19) >DroYak_CAF1 56460 108 + 1 UACUAAACUUUUCCGCAGUCCCCGAUACGGAAUUUCAAUAAUAGAGGACAAUAUGGAACCCCCCAUGUUGUCGAGCUACACAAGACAAUACUCCCAACCGUAUCCAAA .......................(((((((................((((((((((......))))))))))(((...............)))....))))))).... ( -24.86) >DroPer_CAF1 103679 108 + 1 CUCUAAUCCCUCUCGUAGCUCCCGCUAUGAGGCGGUCGACAUUGAGGACGACCUGGAGCUGCCGAUAGUGUCCAGCUACACGCGCCACUACUCCCCGCCGUUUCCCUC ............(((((((....)))))))((((((((.(......).))))).((((.((.((...((((......)))).)).))...))))..)))......... ( -31.10) >consensus UACUUAACUUUUCCGCAGUCCCCGUUACGGAAUUUCCCUAAUCGAGGACAACAUGGAACCCCCCAUGUUGUCGAGCUACACAAGGCAAUACUCCCAGCCGUAUCCAAA .........................(((((................((((((((((......))))))))))(((...............)))....)))))...... (-15.05 = -15.74 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:38 2006