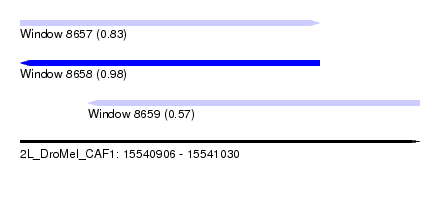
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,540,906 – 15,541,030 |
| Length | 124 |
| Max. P | 0.975951 |

| Location | 15,540,906 – 15,540,999 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
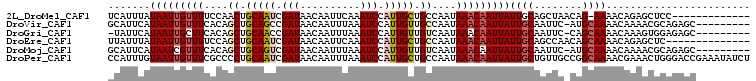
>2L_DroMel_CAF1 15540906 93 + 22407834 -------------GGAGCUCUGUUUU-CUGUUAGCUGCAAUAAUUGUUUAUUGGCAGCAAUGGAUUUGAAUUGUUAUCGAUUGCAGUUGGAAAACAAUUAUAAAUGA -------------(.((((.......-.....)))).).((((((((((.(..((.(((((.(((..........))).))))).))..).))))))))))...... ( -24.40) >DroVir_CAF1 65553 97 + 1 ---------GCUCUGCGUUUUGUUUUGCAU-GAAUUGCAAUAAUUGUUUAUUGGCAACAAUGGAUUUAAAUUGUUAUCGGCUGCAGCUGUGAAACAAUUAUGAAUGC ---------((....(((.((((((..(..-((...(((((....((((((((....))))))))....)))))..))(((....))))..))))))..)))...)) ( -25.00) >DroGri_CAF1 54620 96 + 1 ---------GCUCUCCACUUUGUUUUGCUG-GAAUUGCAAUAAUUGUUUAUUGACAACAAUGGAUUUAAAUUGUUAUCGGUUGCAGCUGUGAAGCAAUUAUGAAUA- ---------......((..(((((((((((-.(((((.(((((((((((((((....))))))))...)))))))..))))).))))...)))))))...))....- ( -20.70) >DroEre_CAF1 48442 93 + 1 --------------GAGCUCUGUUUUGCUGUUGGCUGCAAUAAUUGUUUAUUGGCAGCAAUGGAUUUGAAUUGUUAUCGAUUGCAGCUGGAAAACAAUUAUAAAUAA --------------......(((...((.....)).)))((((((((((.(..((.(((((.(((..........))).))))).))..).))))))))))...... ( -24.90) >DroMoj_CAF1 67845 97 + 1 ---------GCUCUGCGUUUUGUUUUGCAU-GAAUUGCAAUAAUUGUUUAUUGACAACAAUGGAUUUAAAUUGUUAUCGACUGCAGCUGUGAAACGAUUAUGAAUGC ---------.....((((((.....((((.-....))))((((((((((.((((.((((((........)))))).))))..((....)).)))))))))))))))) ( -20.60) >DroPer_CAF1 96267 107 + 1 AGAUAUUUCGGUCCCAGUUUCGUUUUGCCGGCAACAGCAAUAAUUGUUUAUUGGCAGCAAUGGAUUUAAAUUGUUAUCGAUUGCAGGGGCGAAACAAUUACAAAUGG .....(((((.((((..........(((((....)...(((((....)))))))))(((((.(((..........))).))))).)))))))))............. ( -27.10) >consensus _________GCUCUGAGCUUUGUUUUGCUG_GAACUGCAAUAAUUGUUUAUUGGCAACAAUGGAUUUAAAUUGUUAUCGAUUGCAGCUGUGAAACAAUUAUAAAUGA ....................((((((((......(((((((....(((....)))((((((........))))))....)))))))..))))))))........... (-14.58 = -14.08 + -0.50)



| Location | 15,540,906 – 15,540,999 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15540906 93 - 22407834 UCAUUUAUAAUUGUUUUCCAACUGCAAUCGAUAACAAUUCAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCAGCUAACAG-AAAACAGAGCUCC------------- ..........((((((((...........((.......)).........(((((.(((((....)))))))))).....)-)))))))......------------- ( -16.00) >DroVir_CAF1 65553 97 - 1 GCAUUCAUAAUUGUUUCACAGCUGCAGCCGAUAACAAUUUAAAUCCAUUGUUGCCAAUAAACAAUUAUUGCAAUUC-AUGCAAAACAAAACGCAGAGC--------- ((.....(((((((((....((.((((..(((..........)))..)))).))....)))))))))(((((....-.)))))........)).....--------- ( -17.00) >DroGri_CAF1 54620 96 - 1 -UAUUCAUAAUUGCUUCACAGCUGCAACCGAUAACAAUUUAAAUCCAUUGUUGUCAAUAAACAAUUAUUGCAAUUC-CAGCAAAACAAAGUGGAGAGC--------- -............((((((..........(((((((((........)))))))))............((((.....-..))))......))))))...--------- ( -18.90) >DroEre_CAF1 48442 93 - 1 UUAUUUAUAAUUGUUUUCCAGCUGCAAUCGAUAACAAUUCAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCAGCCAACAGCAAAACAGAGCUC-------------- .......(((((((((....((.(((((.(((..........))).))))).))....)))))))))((((........))))..........-------------- ( -18.20) >DroMoj_CAF1 67845 97 - 1 GCAUUCAUAAUCGUUUCACAGCUGCAGUCGAUAACAAUUUAAAUCCAUUGUUGUCAAUAAACAAUUAUUGCAAUUC-AUGCAAAACAAAACGCAGAGC--------- .....................((((....(((((((((........)))))))))............(((((....-.)))))........))))...--------- ( -18.60) >DroPer_CAF1 96267 107 - 1 CCAUUUGUAAUUGUUUCGCCCCUGCAAUCGAUAACAAUUUAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCUGUUGCCGGCAAAACGAAACUGGGACCGAAAUAUCU ...........(((((((.(((.(((((.(((..........))).)))))((((....((((........))))...))))..........)))..)))))))... ( -22.40) >consensus GCAUUCAUAAUUGUUUCACAGCUGCAAUCGAUAACAAUUUAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCAAUUC_CAGCAAAACAAAACGCAGAGC_________ .......(((((((((....((.(((((.(((..........))).))))).))....)))))))))((((........))))........................ (-12.16 = -12.75 + 0.59)
| Location | 15,540,927 – 15,541,030 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -15.45 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.49 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
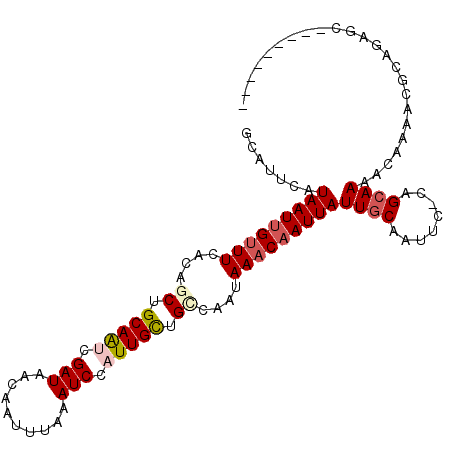
>2L_DroMel_CAF1 15540927 103 - 22407834 CAUAAGUUUAACCCCCGGUAUUCACACUUGGUCAUUUAUAAUUGUUUUCCAACUGCAAUCGAUAACAAUUCAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCA .............((.(((......))).))......((((((((((.......(((((.(((..........))).))))).......)))))))))).... ( -15.24) >DroVir_CAF1 65578 95 - 1 CAUUCGCUUCAUUC-----GUUCGCU---CUGCAUUCAUAAUUGUUUCACAGCUGCAGCCGAUAACAAUUUAAAUCCAUUGUUGCCAAUAAACAAUUAUUGCA ..............-----.......---..(((...((((((((((....((.((((..(((..........)))..)))).))....))))))))))))). ( -14.40) >DroPse_CAF1 97439 97 - 1 CAUGAUUC-GACUC-----GUUCACACUUGGCCAUUUGUAAUUGUUUCGCCCCUGCAAUCGAUAACAAUUUAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCU ........-.....-----..........(((.....((((((((((.......(((((.(((..........))).))))).......)))))))))).))) ( -13.94) >DroEre_CAF1 48463 103 - 1 UUUGAGUUGAACCCCCGGAAUUCACACUUGGUUAUUUAUAAUUGUUUUCCAGCUGCAAUCGAUAACAAUUCAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCA .(..(((((((((...))..)))).)))..)......((((((((((....((.(((((.(((..........))).))))).))....)))))))))).... ( -20.10) >DroYak_CAF1 50619 103 - 1 CAUAAGUUGAGCCCCCGGAAUUCACACUUGAUCAUUUAUAAUUGUUUUCCAGCUGCAAUCGAUAACAAUUCAAAUCCAUUGAUGCCAAUAAACAAUUAUUGCA ..((((((((.((...))...))).))))).......((((((((((....((..((((.(((..........))).))))..))....)))))))))).... ( -15.10) >DroPer_CAF1 96302 97 - 1 CAUGAUUC-GACUC-----GUUCACACUUGGCCAUUUGUAAUUGUUUCGCCCCUGCAAUCGAUAACAAUUUAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCU ........-.....-----..........(((.....((((((((((.......(((((.(((..........))).))))).......)))))))))).))) ( -13.94) >consensus CAUGAGUU_AACCC_____AUUCACACUUGGUCAUUUAUAAUUGUUUCCCAGCUGCAAUCGAUAACAAUUCAAAUCCAUUGCUGCCAAUAAACAAUUAUUGCA .....................................((((((((((.......(((((.(((..........))).))))).......)))))))))).... (-11.63 = -11.49 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:35 2006