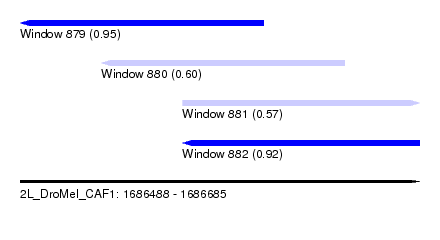
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,686,488 – 1,686,685 |
| Length | 197 |
| Max. P | 0.949058 |

| Location | 1,686,488 – 1,686,608 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1686488 120 - 22407834 CCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGCCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUGUUAAUGCAGUGCAAGUACAAAGAAUGUGUAUCUUGAAGAU ....(((..(((.(((((((((...((((((.((((..(((......)))..))))..))))))...........))))))))))))..)))(((((((.......)))).)))...... ( -37.14) >DroSec_CAF1 5471 120 - 1 CCAAGCGAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUGUUAAUGCAGUGCAAGUACAAACAAUGUGUAUUUUGAAGAU ....(((..(((.(((((((((...((((((.((((..(((......)))..))))..))))))...........))))))))))))..)))(((((((.......)))))))....... ( -35.84) >DroSim_CAF1 6455 120 - 1 CCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGCGCAGCCGAAGAAAAAAUGUUGUGUUAAUGCAGUGCAAGUACAAACAAUGUGUAUCUUGAAGAU ....(((..(((.(((((((((...((((((.((((..(((......)))..))))..))))))...........))))))))))))..)))(((((((.......)))).)))...... ( -34.44) >DroYak_CAF1 6624 105 - 1 CCAAGCAAAGCAGUUAACACAACAA---------------CUAAAACGCUUCGCUGGGCAGCCGAAGAAAAGCUGUUGUGUUGAUGCAGUGCAAGUACAAACAAUGUGUAUCUUGAAGAU ....(((..(((.(((((((((((.---------------((......(((((((....)).)))))...)).))))))))))))))..)))(((((((.......)))).)))...... ( -30.90) >consensus CCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUGUUAAUGCAGUGCAAGUACAAACAAUGUGUAUCUUGAAGAU .((((((..(((.(((((((((....((((........))))......(((((((....)).)))))........))))))))))))..)))..(((((.......))))).)))..... (-26.08 = -26.32 + 0.25)

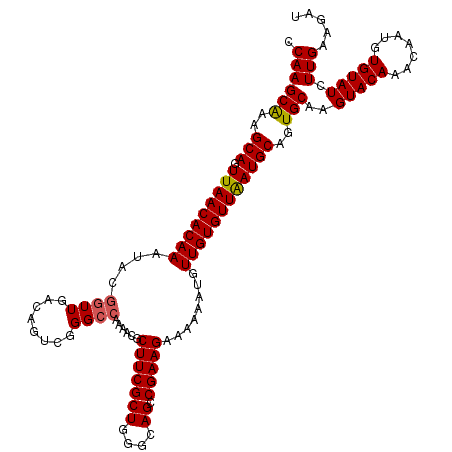
| Location | 1,686,528 – 1,686,648 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -22.31 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1686528 120 - 22407834 UGCAGCGCCAUGAAACACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGCCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUG .((((((((((.........))).((((((........)))..)))...))..............((((((.((((..(((......)))..))))..))))))..........))))). ( -33.50) >DroSec_CAF1 5511 120 - 1 UGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCGAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUG .(((((..(((.........))).)))))........(((((.((((((((.((((((.(......)))))))....(........))))))))).)))..))................. ( -35.40) >DroSim_CAF1 6495 120 - 1 UGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGCGCAGCCGAAGAAAAAAUGUUGUG .((((((((((.........))).((((((........)))..)))...))..............((((((.((((..(((......)))..))))..))))))..........))))). ( -30.80) >DroYak_CAF1 6664 105 - 1 UGCAGCUCCGUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAACAA---------------CUAAAACGCUUCGCUGGGCAGCCGAAGAAAAGCUGUUGUG ((((((((..((.....))...)))))(((........)))...)))..........(((((((.---------------((......(((((((....)).)))))...)).))))))) ( -27.30) >consensus UGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGGCCAAAACGCUUCGCUGGGCAGCCGAAGAAAAAAUGUUGUG .(((((..(((.........))).)))))........(((((.(((.((((.((....))......((((........)))).....)))).))).)))..))................. (-22.31 = -23.00 + 0.69)

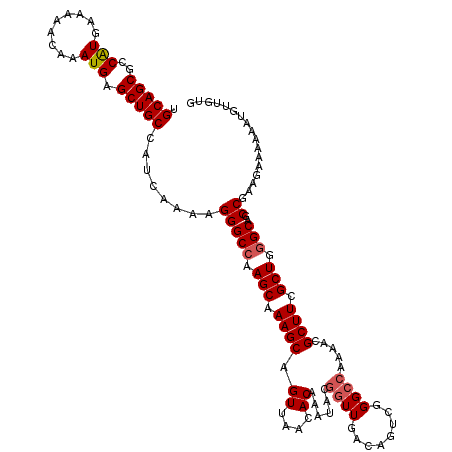
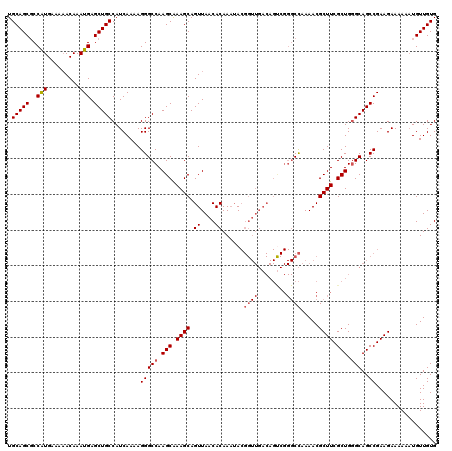
| Location | 1,686,568 – 1,686,685 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -23.69 |
| Energy contribution | -24.44 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1686568 117 + 22407834 CCCGGCUGUCAACCGUAUUUGUGUUAACUGCUUUGCUUGGCCCUUUUGAUGGCAGCUCAUUUGUGUUUCAUGGCGCUGCAUUUCAAGUUGGCUGCACUUUCCUGCAUAGCUAAUUAC--- ...(((.((.(((((....)).))).)).)))..(((.((((..(((((..(((((.(...((.....))..).)))))...)))))..))))(((......)))..))).......--- ( -29.70) >DroSec_CAF1 5551 116 + 1 CCCGACUGUCAACCGUAUUUGUGUUAACUGCUUCGCUUGGCCCUUUUGAUGGCAGCUCAUUUGUUUUUCAUGGCGCUGCAUUUCA-GUUGGCUGCAUAUUCCUGCAUAGCUAAUUAC--- .(((((((((((.((.....((....)).....)).)))))......((..(((((.(...((.....))..).)))))...)))-))))).((((......))))...........--- ( -26.30) >DroSim_CAF1 6535 116 + 1 CCCGACUGUCAACCGUAUUUGUGUUAACUGCUUUGCUUGGCCCUUUUGAUGGCAGCUCAUUUGUUUUUCAUGGCGCUGCAUUUCA-GUUGGCUGCACUUUCCUGCAUAGCUAAUUAC--- .(((((((((((.((.....((....)).....)).)))))......((..(((((.(...((.....))..).)))))...)))-))))).((((......))))...........--- ( -24.50) >DroYak_CAF1 6704 105 + 1 ---------------UUGUUGUGUUAACUGCUUUGCUUGGCCCUUUUGAUGGCAGCUCAUUUGUUUUUCACGGAGCUGCAUUUCAAGUUGGCUGCAUUUUCCUGCAUAGCUAAUUACCAC ---------------..(((((((.........(((..((((..(((((..(((((((...((.....))..)))))))...)))))..))))))).......))))))).......... ( -33.69) >consensus CCCGACUGUCAACCGUAUUUGUGUUAACUGCUUUGCUUGGCCCUUUUGAUGGCAGCUCAUUUGUUUUUCAUGGCGCUGCAUUUCA_GUUGGCUGCACUUUCCUGCAUAGCUAAUUAC___ ..................((((((.........(((..((((..(((((..(((((.(...((.....))..).)))))...)))))..))))))).......))))))........... (-23.69 = -24.44 + 0.75)

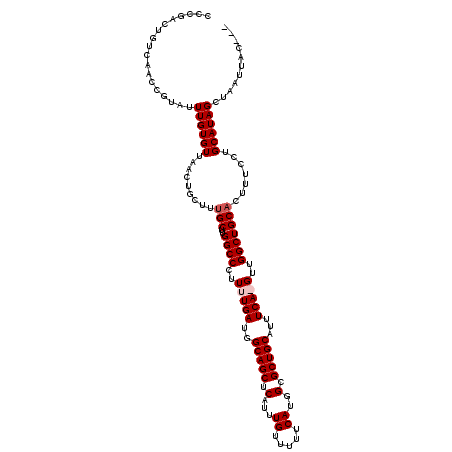
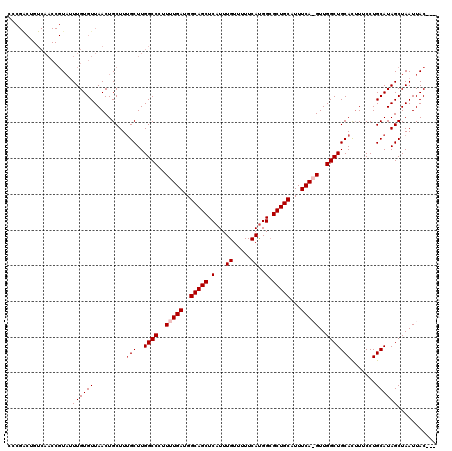
| Location | 1,686,568 – 1,686,685 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1686568 117 - 22407834 ---GUAAUUAGCUAUGCAGGAAAGUGCAGCCAACUUGAAAUGCAGCGCCAUGAAACACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGCCGGG ---.......(((.((((......))))(((...((((...(((((..(((.........))).)))))..))))...)))..)))...((.((((((.(......))))))).)).... ( -29.90) >DroSec_CAF1 5551 116 - 1 ---GUAAUUAGCUAUGCAGGAAUAUGCAGCCAAC-UGAAAUGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCGAAGCAGUUAACACAAAUACGGUUGACAGUCGGG ---.......(((.((((......))))(((...-(((...(((((..(((.........))).)))))..)))....)))..)))......((((((.(......)))))))....... ( -27.80) >DroSim_CAF1 6535 116 - 1 ---GUAAUUAGCUAUGCAGGAAAGUGCAGCCAAC-UGAAAUGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGG ---.......(((.((((......))))(((...-(((...(((((..(((.........))).)))))..)))....)))..)))......((((((.(......)))))))....... ( -26.90) >DroYak_CAF1 6704 105 - 1 GUGGUAAUUAGCUAUGCAGGAAAAUGCAGCCAACUUGAAAUGCAGCUCCGUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAACAA--------------- (((.(((((.(((.((((......))))(((...((((...(((((((..((.....))...)))))))..))))...))).......)))))))).))).....--------------- ( -32.40) >consensus ___GUAAUUAGCUAUGCAGGAAAAUGCAGCCAAC_UGAAAUGCAGCGCCAUGAAAAACAAAUGAGCUGCCAUCAAAAGGGCCAAGCAAAGCAGUUAACACAAAUACGGUUGACAGUCGGG ....(((((.(((.((((......))))(((....(((...(((((..(((.........))).)))))..)))....))).......))))))))........................ (-23.34 = -23.15 + -0.19)

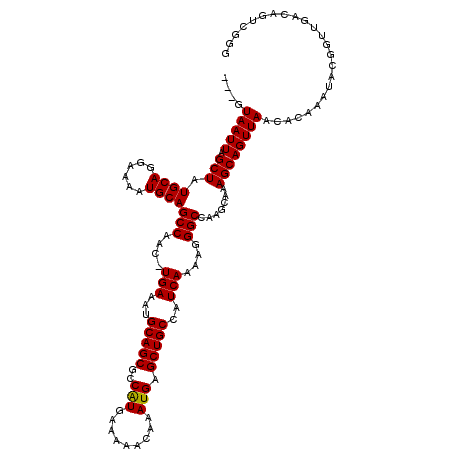
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:59 2006