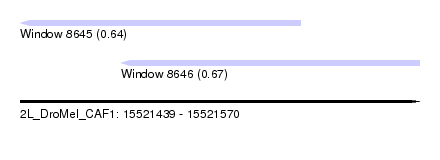
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,521,439 – 15,521,570 |
| Length | 131 |
| Max. P | 0.668025 |

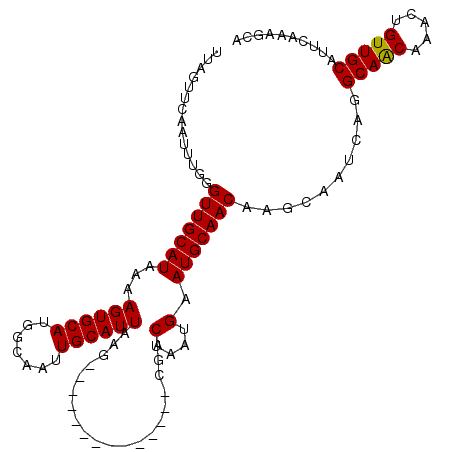
| Location | 15,521,439 – 15,521,531 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -13.01 |
| Energy contribution | -12.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15521439 92 - 22407834 CAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCACUACUAAUGAG-UGGUUGAAUUCAAUUUAAACGA--- ......(--------------((((...(((.(((.....))).)))(....).....)))))((((((..((((.(....)))-)).)))))).............--- ( -19.90) >DroPse_CAF1 73941 106 - 1 CAUUAAGUGCCGAGGAGAUGCCGUCUCGUGAAUGCAAC----AAUCAGGCAGCAAACUGUUGCACUCAAAGCCCUCCUAAUGAACUGGGGGACUACAAUCUAAAUUUAUU .....(((((((((..(....)..))))(((.((((((----(..............))))))).)))..))(((((.........))))))))................ ( -23.34) >DroSim_CAF1 26607 92 - 1 CAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCACUACUAAUGAG-UGGUUGAAUUCAAUUUAGACGA--- .......--------------((.(((((((((.((((..((((.(((........)))))))........((((.(....)))-)))))).)))))..)))).)).--- ( -21.70) >DroEre_CAF1 26522 92 - 1 CAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCACUACUAAUGAG-UGGUUGAAUUCAAUUUAGACGA--- .......--------------((.(((((((((.((((..((((.(((........)))))))........((((.(....)))-)))))).)))))..)))).)).--- ( -21.70) >DroYak_CAF1 26859 92 - 1 CAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCGUUCAAAGCACUACUAAUGAG-UGGUUGAAUUCAAUUUAGACGA--- .......--------------((.(((((((((.((((..((((.(((........)))))))........((((.(....)))-)))))).)))))..)))).)).--- ( -21.70) >DroPer_CAF1 72599 106 - 1 CAUUAAGUGCCGGGGAGAUGCCGUCUCGUGAAUGCAAC----AAUCAGGCAGCAAACUGUUGCACUCAAAGCCCUCCUAAUGAACUGGGGGACUACAAUCUAAAUUUAUU .....(((.((((((((((...))))).(((.((((((----(..............))))))).)))...))).((((......)))))))))................ ( -25.24) >consensus CAUUAAG______________CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCACUACUAAUGAG_UGGUUGAAUUCAAUUUAAACGA___ ......................((....(((((((((((..................)))))))))))...((..((((......))))))...)).............. (-13.01 = -12.20 + -0.80)

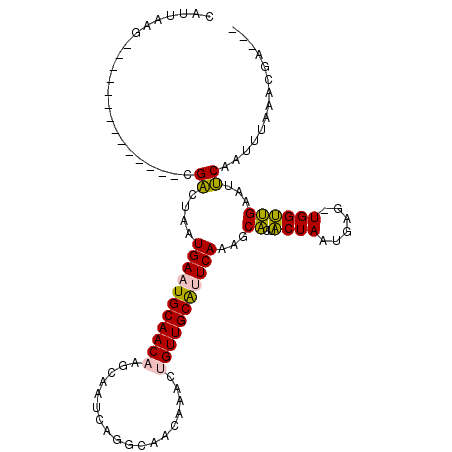
| Location | 15,521,472 – 15,521,570 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.668025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15521472 98 - 22407834 UUAGUUCAAUUUGGGUUGCAUAAAAGUGCAUGGCAAUUGCAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCA ...(((.......((((((.....((((((.......))))))..)--------------)))))..(((((((((((........(....)....))))))))))).))). ( -25.50) >DroPse_CAF1 73978 107 - 1 UUGCCUCGGGCU-UGUUGCAUAAGAGUGCAUGGCAAUUGCAUUAAGUGCCGAGGAGAUGCCGUCUCGUGAAUGCAAC----AAUCAGGCAGCAAACUGUUGCACUCAAAGCC ........((((-((((((((...((((((.......))))))......((((..(....)..))))...)))))))----......(((((.....))))).....))))) ( -31.90) >DroSim_CAF1 26640 98 - 1 UUAGUUCAAUUUGGGUUGCAUAAAAGUGCAUGGCAAUUGCAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCA ...(((.......((((((.....((((((.......))))))..)--------------)))))..(((((((((((........(....)....))))))))))).))). ( -25.50) >DroEre_CAF1 26555 98 - 1 UUAGUUCAAUUUGGGUUGCAUAAAAGUGCAUGGCAAUUGCAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCA ...(((.......((((((.....((((((.......))))))..)--------------)))))..(((((((((((........(....)....))))))))))).))). ( -25.50) >DroYak_CAF1 26892 98 - 1 UUAGUUCAAUUUGGGUUGCAUAAAAGUGCAUGGCAAUUGCAUUAAG--------------CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCGUUCAAAGCA ...(((.......((((((.....((((((.......))))))..)--------------)))))..(((((((((((........(....)....))))))))))).))). ( -25.10) >DroPer_CAF1 72636 107 - 1 UUGCCUCGGGCU-UGUUGCAUAAGAGUGCAUGGCAAUUGCAUUAAGUGCCGGGGAGAUGCCGUCUCGUGAAUGCAAC----AAUCAGGCAGCAAACUGUUGCACUCAAAGCC ........((((-((((((((..(((.((((..(..(.((((...)))).)..)..))))...)))....)))))))----......(((((.....))))).....))))) ( -31.80) >consensus UUAGUUCAAUUUGGGUUGCAUAAAAGUGCAUGGCAAUUGCAUUAAG______________CGACUAAUGAAUGCAACAAGCAAUCAGGCAACAAACUGUUGCAUUCAAAGCA ..............(((((((...((((((.......))))))....................(....).)))))))..........(((((.....))))).......... (-18.75 = -18.53 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:22 2006