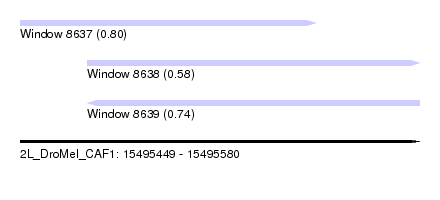
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,495,449 – 15,495,580 |
| Length | 131 |
| Max. P | 0.800764 |

| Location | 15,495,449 – 15,495,546 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15495449 97 + 22407834 CAAAAAU---GUGGUUCCAUAGCUGGUUCCAUUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAAUUAUUGAUUUUGUGUGAGAGCGAGGGAG--CGCGCAUA .....((---((((((((......(((.......)))..((((((((((.....)))..(((((........)))))....)))))))))))--).))))). ( -30.30) >DroEre_CAF1 260 94 + 1 CAAAAAUCGCGUGGUGCCACUGCUCG---CGCUCUCCCUCUCGCUCGCUCCUCCAGCCGGCGAACCAUUGAUUUUGCGCGAGAGCGUGUGAG--UGCGA--- ........(((((....))).))(((---(((((.(((((((((..(((.....)))..(((((........)))))))))))).).).)))--)))))--- ( -36.70) >DroYak_CAF1 304 93 + 1 CUAAAAU---GUGGUUCCAUUGCUCG---CGCUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAACCAUUCAUUUUGUGGGAGAGCGAGCGAGAGUGAGA--- .......---(((((......)).))---).(((((..((((((((((((((((.....(((((........)))))))))))))))))))))))))).--- ( -41.70) >consensus CAAAAAU___GUGGUUCCAUUGCUCG___CGCUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAACCAUUGAUUUUGUGCGAGAGCGAGCGAG__UGCGA___ ..........(((....)))...................(((((((((((((...((..(((((........)))))))))))))))))))).......... (-20.69 = -21.37 + 0.67)

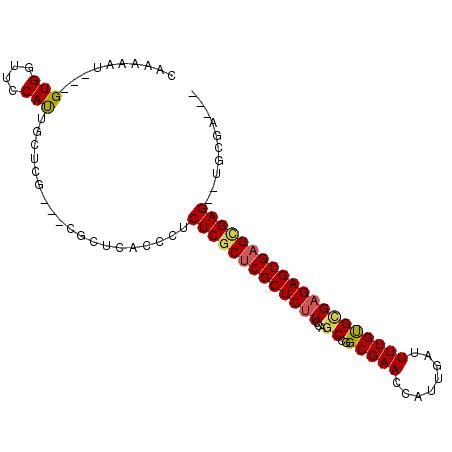
| Location | 15,495,471 – 15,495,580 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.04 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.45 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15495471 109 + 22407834 GUUCCAUUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAAUUAUUGAUUUUGUGUGAGAGCGAGGGAG--CGCGCAUAAGAGAGAGGGAGAGAGAGUGAGAGUGAAGGCGUG ...((.(((((..((((((((.(((((((.(....)......((..(((((((((...((......)--).)))))))))..))))))))).))))))))))))))).... ( -42.80) >DroEre_CAF1 285 90 + 1 G---CGCUCUCCCUCUCGCUCGCUCCUCCAGCCGGCGAACCAUUGAUUUUGCGCGAGAGCGUGUGAG--UGCGA------GAGAG------A----GAGCGAGAGGGCGUG (---(((((((((((((.(((((..(((((....(((((........)))))((....)).)).)))--.))))------)...)------)----))).).)))))))). ( -42.00) >DroYak_CAF1 326 91 + 1 G---CGCUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAACCAUUCAUUUUGUGGGAGAGCGAGCGAGAGUGAGA------GAGAG------C----GAGCGAGAG-GCGUG .---....((((((((((((((((((..(..((.(((((........))))).)).......((....))..).------.))))------)----)))))))))-).))) ( -45.10) >DroMoj_CAF1 374 94 + 1 -------UCUGCUUCUUGUUCU--CCUCGUGCCGGUGCUUCAUUGAUUUUGCUUGCAAGUAUGUGU--GUGUGCGA----GAGAG--CAAGUCCAUGAAUAAGCAGGCGUG -------(((((((((((((((--(.(((..(((((((((((...........)).))))))...)--).)..)))----)))))--)))).........))))))).... ( -29.70) >consensus G___CGCUCACCCUCUCGCUCGCUCCUCCAGCCGGCGAACCAUUGAUUUUGCGUGAGAGCGAGUGAG__UGCGA______GAGAG______A____GAGCGAGAAGGCGUG ........(.(((((((((((............((....))......(((((......))))).................................))))))))))).).. (-14.64 = -14.45 + -0.19)

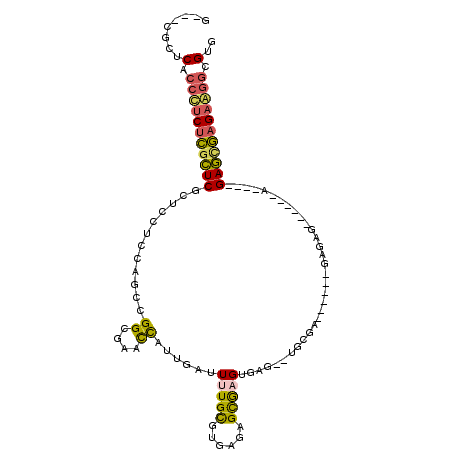

| Location | 15,495,471 – 15,495,580 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 63.04 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -13.39 |
| Energy contribution | -15.70 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15495471 109 - 22407834 CACGCCUUCACUCUCACUCUCUCUCCCUCUCUCUUAUGCGCG--CUCCCUCGCUCUCACACAAAAUCAAUAAUUCGCCGGCUGGAAGAGCGAGCGAGAGGGUGAAUGGAAC ......(((.(..((((((((((...(((.(((((....(((--......))).......................((....))))))).))).))))))))))..)))). ( -29.00) >DroEre_CAF1 285 90 - 1 CACGCCCUCUCGCUC----U------CUCUC------UCGCA--CUCACACGCUCUCGCGCAAAAUCAAUGGUUCGCCGGCUGGAGGAGCGAGCGAGAGGGAGAGCG---C ..(.(((((((((((----.------(((.(------((.((--..(...(((....)))..........((....)))..)))))))).))))))))))).)....---. ( -39.10) >DroYak_CAF1 326 91 - 1 CACGC-CUCUCGCUC----G------CUCUC------UCUCACUCUCGCUCGCUCUCCCACAAAAUGAAUGGUUCGCCGGCUGGAAGAGCGAGCGAGAGGGUGAGCG---C .....-....(((((----(------((...------.....((((((((((((((.(((.........(((....)))..))).))))))))))))))))))))))---. ( -43.00) >DroMoj_CAF1 374 94 - 1 CACGCCUGCUUAUUCAUGGACUUG--CUCUC----UCGCACAC--ACACAUACUUGCAAGCAAAAUCAAUGAAGCACCGGCACGAGG--AGAACAAGAAGCAGA------- .....((((((.((....))((((--.((((----(((.....--........(((....)))..........((....)).))).)--))).)))))))))).------- ( -18.90) >consensus CACGCCCUCUCGCUC____U______CUCUC______GCGCA__CUCACACGCUCUCACACAAAAUCAAUGAUUCGCCGGCUGGAAGAGCGAGCGAGAGGGAGAGCG___C ..(((((((((((((......................................................(((....)))(((.....))))))))))))))))........ (-13.39 = -15.70 + 2.31)

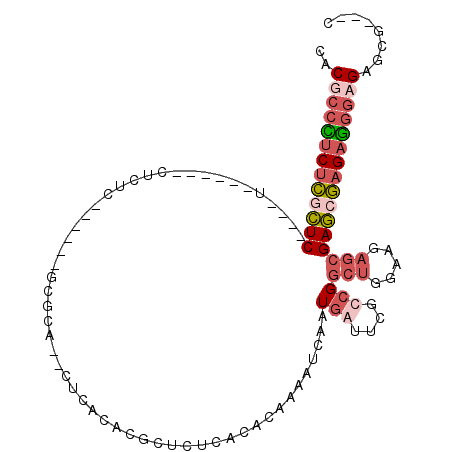
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:16 2006