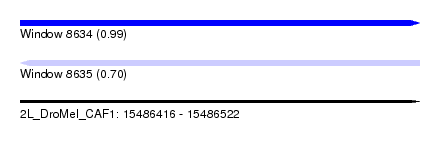
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,486,416 – 15,486,522 |
| Length | 106 |
| Max. P | 0.990426 |

| Location | 15,486,416 – 15,486,522 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -33.62 |
| Energy contribution | -34.46 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
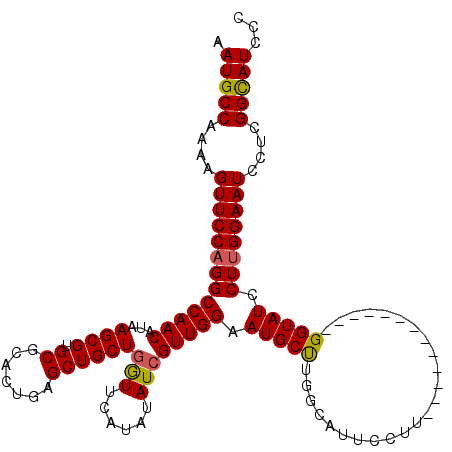
>2L_DroMel_CAF1 15486416 106 + 22407834 GGGAUGCCGAGGAUUCCAAGGAUACC-------------ACGGAAUACCAAGCAUUCCAACGAUAUAUGAACCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU ..(((((((((..(((((.((.....-------------..(((((.......))))).............((((((....((((....))))..)))))))))))))..))))))))) ( -34.10) >DroSec_CAF1 225095 106 + 1 GGGAUGCCGAGGAUUCCAAGGAUACC-------------AAGGAAUGCCAAGCAUUCCAACGAUAUAUGAACCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU ..(((((((((..(((((.((.....-------------..(((((((...))))))).............((((((....((((....))))..)))))))))))))..))))))))) ( -39.40) >DroSim_CAF1 231492 106 + 1 GGGAUGCCGAGGAUUCCAAGGAUACC-------------AAGGAAUGCCAAGCAUUCCAACGAUAUAUGAACCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU ..(((((((((..(((((.((.....-------------..(((((((...))))))).............((((((....((((....))))..)))))))))))))..))))))))) ( -39.40) >DroEre_CAF1 234947 105 + 1 GGGAUGCCAAGGAUUCCAAGGAUACC-------------GAGG-CAGCCUGGCAUUCCAACCAUAUAGGAACCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU ..(((((((((..((((..((...))-------------.(((-(...((((..((((.........))))))))((((..((((....))))...))))))))))))..))))))))) ( -35.80) >DroYak_CAF1 234132 118 + 1 GGGAUACCACGGAUUCCGAGGAUACCAACCAAGGCUUCCAAGG-AUGCCAGGCAUUCCAACGAUACUUGAAUCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU (((((.(....))))))..(((..((......))..)))...(-((((((((..(((((..(((......)))..((((..((((....))))...))))...)))))..))))))))) ( -34.10) >consensus GGGAUGCCGAGGAUUCCAAGGAUACC_____________AAGGAAUGCCAAGCAUUCCAACGAUAUAUGAACCAGCAGCUCAGUGCGCACGCUUAUGUUGGCCUGGAACUUUUGGCAUU ..(((((((((..(((((.((....................(((((((...))))))).............((((((....((((....))))..)))))))))))))..))))))))) (-33.62 = -34.46 + 0.84)



| Location | 15,486,416 – 15,486,522 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15486416 106 - 22407834 AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGGUUCAUAUAUCGUUGGAAUGCUUGGUAUUCCGU-------------GGUAUCCUUGGAAUCCUCGGCAUCCC .(((((....(((((((((((((....((....))..(((((.....)))))......))))).((((((((....))).-------------))))).))))))))....)))))... ( -32.50) >DroSec_CAF1 225095 106 - 1 AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGGUUCAUAUAUCGUUGGAAUGCUUGGCAUUCCUU-------------GGUAUCCUUGGAAUCCUCGGCAUCCC .(((((....((((((((..(((...((((.((.......)))))).)))...((((((..(((((((...))))))).)-------------))))).))))))))....)))))... ( -34.40) >DroSim_CAF1 231492 106 - 1 AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGGUUCAUAUAUCGUUGGAAUGCUUGGCAUUCCUU-------------GGUAUCCUUGGAAUCCUCGGCAUCCC .(((((....((((((((..(((...((((.((.......)))))).)))...((((((..(((((((...))))))).)-------------))))).))))))))....)))))... ( -34.40) >DroEre_CAF1 234947 105 - 1 AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGGUUCCUAUAUGGUUGGAAUGCCAGGCUG-CCUC-------------GGUAUCCUUGGAAUCCUUGGCAUCCC .(((((((..((((((((.........((....))(((((((.(((((((((.........)))..)))).)).)-.)))-------------)))...))))))))..)))))))... ( -38.20) >DroYak_CAF1 234132 118 - 1 AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGAUUCAAGUAUCGUUGGAAUGCCUGGCAU-CCUUGGAAGCCUUGGUUGGUAUCCUCGGAAUCCGUGGUAUCCC .((((((...(((((((((((((...((((.((.......))))))(((......)))))))).(((((.(((.(-(....)).)))......)))))))).)))))...))))))... ( -38.20) >consensus AAUGCCAAAAGUUCCAGGCCAACAUAAGCGUGCGCACUGAGCUGCUGGUUCAUAUAUCGUUGGAAUGCUUGGCAUUCCUU_____________GGUAUCCUUGGAAUCCUCGGCAUCCC .(((((....(((((((((((((...((((.((.......))))))(((......)))))))).(((((........................))))).))))))))....)))))... (-27.06 = -26.90 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:12 2006