
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,483,859 – 15,483,949 |
| Length | 90 |
| Max. P | 0.956969 |

| Location | 15,483,859 – 15,483,949 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15483859 90 + 22407834 U--------U-UCAACCGCAUCGAUAAUCGCUU-UCAAUUUUCUACUGCCGCUUAGUAUGCAUGCGG-------UCC--------AUUUGCAUGUUUUCGGCUGAUGAAAUUGAU .--------.-((((...((((((.........-))...........((((........((((((((-------...--------..))))))))...)))).))))...)))). ( -19.70) >DroVir_CAF1 260193 99 + 1 --------------AUCGCAUCGAUAAUUGUUUCUCAAUUUUCGGCUACCGCUUCAUAUGCAUGCAAUGUGCGAUCCCAAAGCUAAUUUGCGU--UUUUGGCUGAUGAAACCGAU --------------((((((((((.(((((.....))))).))(((((..((.......))((((((...((.........))....))))))--...)))))))))....)))) ( -20.80) >DroEre_CAF1 232377 90 + 1 U--------U-UCAACCGCAUCGAUAAUCGCUU-UCAAUUUUCUACUGCCGCUUAGUAUGCAUGCGA-------UCC--------AUUUGCAUGUUUUCGGCUGAUGAAAUUGAU .--------.-((((...((((((.........-))...........((((........((((((((-------...--------..))))))))...)))).))))...)))). ( -20.50) >DroYak_CAF1 231438 90 + 1 U--------U-UCAACCGCAUCGAUAAUCGCUU-UCAAUUUUCUACUGCCGCUUAGUAUGCAUGCGA-------UCC--------AUUUGCAUGUUUUCGGCUGAUGAAAUUGAU .--------.-((((...((((((.........-))...........((((........((((((((-------...--------..))))))))...)))).))))...)))). ( -20.50) >DroMoj_CAF1 272700 114 + 1 CUGAUGUGAG-UCGAUCGCAUCGAUAAUCGUUUCUCAAUUUUCGACUGCCGCUUCAUAUGCAUGCAAUGUGCCAUCCGAAAGCUAAUUUGCGUAUUUUUGGCUGAUGAAAACAAU .((((((((.-....))))))))...............((((((.(.((((....(((((((.((.....))....(....)......)))))))...)))).).)))))).... ( -25.60) >DroAna_CAF1 238704 92 + 1 U--------UUUGAACCGCAUCGAUGAUCGCUU-UCAAUUUCCAACUGCCGCUUAGUAUGCAUGCGG------AACC--------AUUUGCAUAUUUUCGGCUGAUGAAAUUGAU .--------........(((((...))).))..-((((((((.....((((...((((((((...(.------...)--------...))))))))..))))....)))))))). ( -23.90) >consensus U________U_UCAACCGCAUCGAUAAUCGCUU_UCAAUUUUCUACUGCCGCUUAGUAUGCAUGCGA_______UCC________AUUUGCAUGUUUUCGGCUGAUGAAAUUGAU ..................................((((((((.....((((...((((((((..........................))))))))..))))....)))))))). (-13.24 = -14.05 + 0.81)
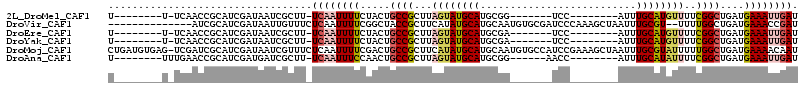

| Location | 15,483,859 – 15,483,949 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15483859 90 - 22407834 AUCAAUUUCAUCAGCCGAAAACAUGCAAAU--------GGA-------CCGCAUGCAUACUAAGCGGCAGUAGAAAAUUGA-AAGCGAUUAUCGAUGCGGUUGA-A--------A .(((((..((((.((((.....(((((..(--------(..-------...)))))))......))))....((.(((((.-...))))).))))))..)))))-.--------. ( -24.40) >DroVir_CAF1 260193 99 - 1 AUCGGUUUCAUCAGCCAAAA--ACGCAAAUUAGCUUUGGGAUCGCACAUUGCAUGCAUAUGAAGCGGUAGCCGAAAAUUGAGAAACAAUUAUCGAUGCGAU-------------- ...((((.....))))....--.((((.......(((((.(((((.(((((....)).)))..)))))..)))))(((((.....))))).....))))..-------------- ( -20.70) >DroEre_CAF1 232377 90 - 1 AUCAAUUUCAUCAGCCGAAAACAUGCAAAU--------GGA-------UCGCAUGCAUACUAAGCGGCAGUAGAAAAUUGA-AAGCGAUUAUCGAUGCGGUUGA-A--------A .(((((..((((.((((.....(((((..(--------(..-------...)))))))......))))....((.(((((.-...))))).))))))..)))))-.--------. ( -24.40) >DroYak_CAF1 231438 90 - 1 AUCAAUUUCAUCAGCCGAAAACAUGCAAAU--------GGA-------UCGCAUGCAUACUAAGCGGCAGUAGAAAAUUGA-AAGCGAUUAUCGAUGCGGUUGA-A--------A .(((((..((((.((((.....(((((..(--------(..-------...)))))))......))))....((.(((((.-...))))).))))))..)))))-.--------. ( -24.40) >DroMoj_CAF1 272700 114 - 1 AUUGUUUUCAUCAGCCAAAAAUACGCAAAUUAGCUUUCGGAUGGCACAUUGCAUGCAUAUGAAGCGGCAGUCGAAAAUUGAGAAACGAUUAUCGAUGCGAUCGA-CUCACAUCAG ((((((((..(((((((.......((......)).......))))..(((((.(((.......)))))))).......)))))))))))....((((.((....-.)).)))).. ( -23.14) >DroAna_CAF1 238704 92 - 1 AUCAAUUUCAUCAGCCGAAAAUAUGCAAAU--------GGUU------CCGCAUGCAUACUAAGCGGCAGUUGGAAAUUGA-AAGCGAUCAUCGAUGCGGUUCAAA--------A .((((((((..(.((((....((((((..(--------(...------.))..)))))).....)))).)...))))))))-..((.(((...)))))........--------. ( -23.90) >consensus AUCAAUUUCAUCAGCCGAAAACAUGCAAAU________GGA_______UCGCAUGCAUACUAAGCGGCAGUAGAAAAUUGA_AAGCGAUUAUCGAUGCGGUUGA_A________A .(((((..((((.((((.....(((((..........................)))))......))))....((.(((((.....))))).))))))..)))))........... (-16.29 = -16.90 + 0.61)

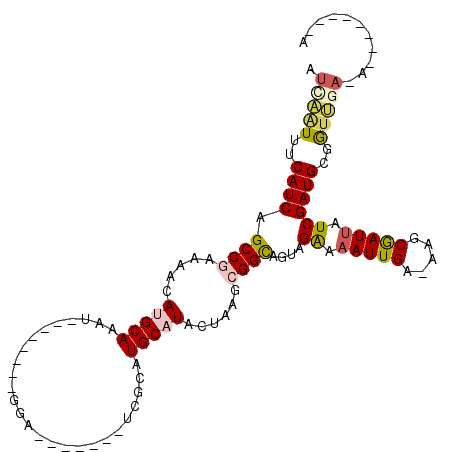
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:10 2006