| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,483,216 – 15,483,327 |
| Length | 111 |
| Max. P | 0.901699 |

| Location | 15,483,216 – 15,483,327 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -21.11 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
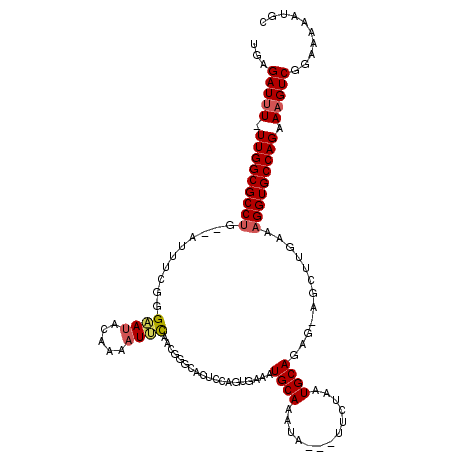
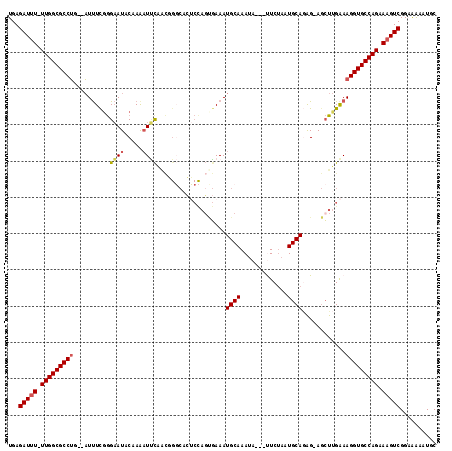
>2L_DroMel_CAF1 15483216 111 - 22407834 UGAGAUUU-UUGGCGCCUG--AUUUUGGGAAUACCAAAUUCAACGGGCACUCCAGUGAAAUGCAAAUA---UUCUAAUGCAUAG-AGCUUGAAAGGUGCCAGAAAGUCGGAAAUAUGC .((..(((-((((((((((--(.(((((.....))))).))..(((((((....))...(((((....---......)))))..-.)))))..)))))))))))).)).......... ( -34.50) >DroVir_CAF1 259654 103 - 1 UGAGAUAU-UUGGCGCCGA--CUCUCGAGCAUACGAAAUGCG------------GAUAAAUGCAAACACAAUUCUAAUGCAAGGCACAUCGAAAGGUGCCAGCAAGUCGGCGAAAUUC ........-....((((((--((..(..((((..(((.((.(------------............).)).)))..))))..(((((..(....)))))).)..))))))))...... ( -30.60) >DroSec_CAF1 221063 111 - 1 UGAGAUUU-UUGGCGCCUG--AUUUCGGGAAUACAAGUUUCAACGGGCACUCCAGUGAAAUGCAAAUA---UUCAAAUGCAGAG-AGCUUGAAAGGUGCCAGAAAGUCGGAAAUAUGC .((..(((-(((((((((.--......(((((....)))))..(((((.(((........((((....---......)))))))-.)))))..)))))))))))).)).......... ( -30.50) >DroSim_CAF1 228307 111 - 1 UGAGAUUU-UUGGCGCCUG--AUUUCGGGAAUACAAAAUUCAACGGGCACUCCAGUGAAAUGCAAAUA---UUCUAAUGCAGAG-AGCUUGAAAGGUGCCAGAAAGUCGGAAAUAUGC .((..(((-(((((((((.--.......((((.....))))..(((((.(((........((((....---......)))))))-.)))))..)))))))))))).)).......... ( -29.80) >DroYak_CAF1 230757 111 - 1 UGAGAUUU-UUGGCGCCUG--AUUUCGGGAAUACAAAAUUCAACUGGCACUCCAGCGAAAUGCAAAUC---CUCUAAUGCAGAG-AGCUUGAAAGGUGCCAGAAAGUCGGAAAAAUGC .....(((-(((((.((((--....))))..............(((((((((.(((....((((....---......))))...-.))).))...)))))))...))))))))..... ( -32.00) >DroAna_CAF1 238039 114 - 1 UGAGAUUUUUUGGCGCCUGGGAUUUCUGGAAUACGAAAUCUAGUUGUCCAGCUAUCGAAAUGCAAAUC---CUCUAAUGCAGAG-AGCUUGGAAGGUGCCAGCAAGUCGGUAGAAUUC ...(((((.(((((((((.(((((((........))))))).....(((((((.((....((((....---......)))))).-))).))))))))))))).))))).......... ( -36.40) >consensus UGAGAUUU_UUGGCGCCUG__AUUUCGGGAAUACAAAAUUCAACGGGCACUCCAGUGAAAUGCAAAUA___UUCUAAUGCAGAG_AGCUUGAAAGGUGCCAGAAAGUCGGAAAAAUGC ...(((((.(((((((((..........((((.....))))...................((((.............))))............))))))))).))))).......... (-21.11 = -21.42 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:08 2006