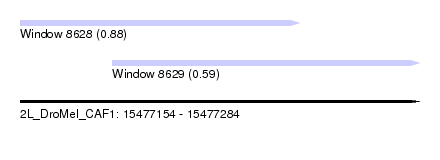
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,477,154 – 15,477,284 |
| Length | 130 |
| Max. P | 0.875653 |

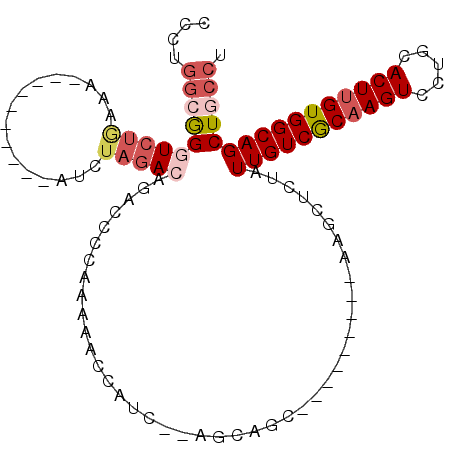
| Location | 15,477,154 – 15,477,245 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -16.94 |
| Energy contribution | -18.22 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15477154 91 + 22407834 CCCUGGCGGGUCUGAAA----------AUCUAAACGGACCCCAAAAACCAUCCCAGCAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ....((((((((((...----------.......))))))).............(((((.---------...))....((((((((((.....)))))))))))))))). ( -29.80) >DroSec_CAF1 215100 91 + 1 CCGUGGCGGGUCUGAAA----------AUCUAGACGGACCCCAAAAACCAUCCCAGCAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ..((((.(((((((...----------.......)))))))......))))....(((((---------.........((((((((((.....))))))))))))))).. ( -29.80) >DroSim_CAF1 222354 79 + 1 CCCUGGCGGGUCUGAAA----------AUCUAGACGGACCCCAAAA------------GC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ....((((((((((...----------.......))))))).....------------..---------.((((.....(((((((((.....)))))))))))))))). ( -28.70) >DroEre_CAF1 225828 79 + 1 CCCUGGAGGAUCUAAAA----------AUCUAGACAGACCCC------------AACAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ...(((.((.((((...----------...))))....))))------------)...((---------(.(((.....(((((((((.....))))))))))))))).. ( -21.00) >DroYak_CAF1 224256 91 + 1 CUCCGGCGGGUCUGAAA----------AUCUAGACAGAGCCCCAAAUCCAUCGGAGCAGC---------AAGCUCUUUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ....((((((((((...----------...)))))(((((......(((...))).....---------..))))).(((((((((((.....)))))))))))))))). ( -32.32) >DroAna_CAF1 231144 103 + 1 -------AGAUCCAAAAAAAGACUUUGAUCCAGACAGGACCCCAAAACGAGAAAAGAAGAGGAGAAGUUGUGCUCUAUUGUCACCAGUCCUGCACUUGUGGCAGCUGCCU -------.((((.(((.......)))))))..(.((((((......((((..........((((........)))).)))).....)))))))......(((....))). ( -16.80) >consensus CCCUGGCGGGUCUGAAA__________AUCUAGACAGACCCCAAAAACCAUC__AGCAGC_________AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCU ....((((((((((................)))))..........................................(((((((((((.....)))))))))))))))). (-16.94 = -18.22 + 1.28)

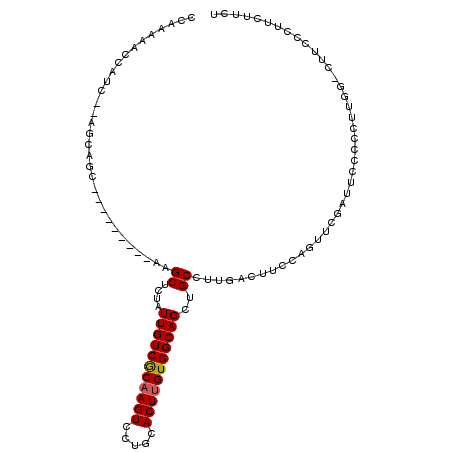
| Location | 15,477,184 – 15,477,284 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15477184 100 + 22407834 CCAAAAACCAUCCCAGCAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACUUCCAGUUCGAUUCCCCCUUGG-CUUCCCUUCUUCU ((((...........(((((---------.........((((((((((.....))))))))))))))).((((........)))).......))))-............. ( -20.90) >DroSec_CAF1 215130 100 + 1 CCAAAAACCAUCCCAGCAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACCUUCAGUUCGAUUCCCCCUUGG-CUUCCCUUCUUCU ((((...........(((((---------.........((((((((((.....))))))))))))))).((((.(....).)))).......))))-............. ( -21.30) >DroSim_CAF1 222384 88 + 1 CCAAAA------------GC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACUUCCAGUUCGAUCCCCCCUUGG-CUUCCCUUCUUCU ....((------------((---------(((..(..(((((((((((.....)))))))))))..).)))).))).......((..((.....))-..))......... ( -18.10) >DroEre_CAF1 225858 76 + 1 CC------------AACAGC---------AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACUUCCAGUUCGAUUCCCC-------------UUUUCU ..------------...(((---------(((..(..(((((((((((.....)))))))))))..).)))).))................-------------...... ( -16.00) >DroYak_CAF1 224286 99 + 1 CCCAAAUCCAUCGGAGCAGC---------AAGCUCUUUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACUUCCAGUUCGAUUCCCCC-UGG-AUUCCCUUUUCCU ....((((((..((((((((---------.........((((((((((.....))))))))))))))).((((........)))).)))...-)))-))).......... ( -26.70) >DroAna_CAF1 231177 102 + 1 CCCAAAACGAGAAAAGAAGAGGAGAAGUUGUGCUCUAUUGUCACCAGUCCUGCACUUGUGGCAGCUGCCUUGACUCCGUCCUGGAUUCCCCCUCAGGAUU--------CU .........((((.......((((.....((((...((((....))))...))))....(((....)))....)))).((((((........))))))))--------)) ( -25.10) >consensus CCAAAAACCAUC__AGCAGC_________AAGCUCUAUUGUCGCAAGUCCUGCACUUGUGGCAGCUGCCUUGACUUCCAGUUCGAUUCCCCCUUGG_CUUCCCUUCUUCU ...............................((....(((((((((((.....)))))))))))..)).......................................... (-13.31 = -13.33 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:06 2006