| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,473,427 – 15,473,536 |
| Length | 109 |
| Max. P | 0.805197 |

| Location | 15,473,427 – 15,473,536 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -28.15 |
| Energy contribution | -26.63 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15473427 109 + 22407834 AAGGUGGAACAGAGCCGGCAGUGGGAUGUCAUUUCGCACUGAAAGAUCCUCUGGCUCGGAACCGGUUUCC--AGAUCGGAUCCGGGGCUGGAGGGCGGCGUCA---UC------UGCUGG ..((..((.....((((.(((((.((.......)).))))).....(((((..(((((((.((((((...--.)))))).))).))))..)))))))))....---))------..)).. ( -50.90) >DroVir_CAF1 248282 103 + 1 GAGGUGGAACAGCGCUGGCAGCGGAAUGUCGUUGCGCACGGAGAGAUCCUCCGGCUCGGAGCUUGC--------AUCCGAGCCAGGGCUCGAAGGUGGCGUCA---UC------UGCUGA ..((((......))))..(((((((.((.(((..(....((((.....))))((((((((......--------.))))))))...........)..))).))---))------))))). ( -45.50) >DroGri_CAF1 229654 103 + 1 GAGAUGGAACAGUGCCGGCAGCGGAAUGUCAUUGCGCACGGAGAGAUCCUCCGGUUCGGAGCUGGC--------AUCCGAGCCGGGACUUGAUGGUGGCGUCA---UC------UGCUGA .((((((.(((((.(((((..((((.(((((((.((..(((((.....)))))...)).)).))))--------))))).))))).))).....))....)))---))------)..... ( -40.70) >DroWil_CAF1 345875 112 + 1 GAGGUGGAAUAAAGCUGGCAAUGGUAUAUCAUUACGCACUGAGAGAUCCUCGGGUUCCGA-----UAGACAAAUAUCGGAGCCAGGACUCGAUGGCGGUGUCA---UCAUUGAUUGCUGA ............(((...((((((((......)))((((((.(((.((((..((((((((-----((......))))))))))))))))).....))))))..---.)))))...))).. ( -40.50) >DroMoj_CAF1 259657 103 + 1 GAGGUGGAACAGCGCUGGGAGCGGAAUGUCGUUGCGCACCGAGAGAUCCUCCGGCUCGGAGCUGGC--------AUCCGAGCCAGGGCUCGAGGGCGGCGUCA---UC------UGCUGG ..((((......))))...((((((.((.((((((...((..(((.((((..((((((((......--------.)))))))))))))))..)))))))).))---))------)))).. ( -46.90) >DroAna_CAF1 226839 112 + 1 GAGGUGGAACAGAGCCGGCAGUGGAAUGUCGUUGCGCACGGAGAGAUCCUCGGGUUCCGAGCCGGAGUCC--ACCUCGGAACCGGGACUCGAUGGCGGUGUCAUCAUC------UGCUGG ((((((((......(((((.(((.(((...))).))).((((...(((....))))))).)))))..)))--)))))....((((.(...((((((...))))))...------).)))) ( -44.50) >consensus GAGGUGGAACAGAGCCGGCAGCGGAAUGUCAUUGCGCACGGAGAGAUCCUCCGGCUCGGAGCUGGC________AUCCGAGCCAGGACUCGAUGGCGGCGUCA___UC______UGCUGA .............((((.(.(((.(((...))).))).....(((.((((..((((((((...............)))))))))))))))...).))))..................... (-28.15 = -26.63 + -1.52)

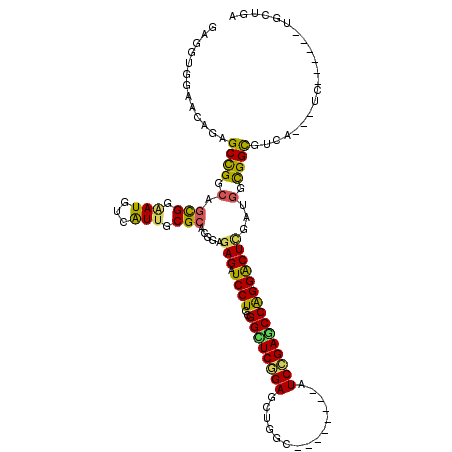

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:03 2006