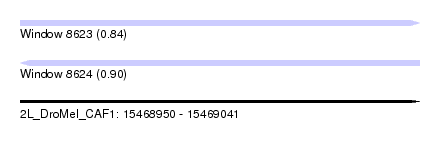
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,468,950 – 15,469,041 |
| Length | 91 |
| Max. P | 0.897366 |

| Location | 15,468,950 – 15,469,041 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -15.78 |
| Energy contribution | -17.28 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15468950 91 + 22407834 ----------CCAUCCCGAUCCCUAUCCCUACUC--CGAUCUCGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ----------....(((((((..........(((--.(....))))..((((((....((.........))((....))...........))))))))))))) ( -23.60) >DroGri_CAF1 224554 73 + 1 ----------CCAGCGCGUC-------------------CCUGACGAUGAUGAGCAUAGGAAACACAAAACGACAUGUCAACUAUAACCACCCAUCGAUCG-A ----------.(((......-------------------.))).((((((((......(....).......((....)).............)))).))))-. ( -10.60) >DroSec_CAF1 206987 93 + 1 ----------CCAUCCCGAUCCCAAUCCCUACCCUACAAUCUGGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ----------....(((((((...(((((.............)).)))((((((....((.........))((....))...........))))))))))))) ( -24.42) >DroSim_CAF1 213078 91 + 1 ----------CCAUCCCGAUCCCAAUCCCUACUC--CGAUCUGGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ----------....(((((((..........(((--(.....))))..((((((....((.........))((....))...........))))))))))))) ( -27.30) >DroEre_CAF1 217645 95 + 1 AUCC------CCAUCCCGAUCCCGAUCCCCACUC--UGAUCUCGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ....------....(((((((..((((.......--.)))).......((((((....((.........))((....))...........))))))))))))) ( -25.00) >DroYak_CAF1 215163 101 + 1 AUCCCCAUCCCCAUCCCGAUCCCAAUCCCCACUC--CGAUCCCGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ..............(((((((..........(((--.......)))..((((((....((.........))((....))...........))))))))))))) ( -23.50) >consensus __________CCAUCCCGAUCCCAAUCCCUACUC__CGAUCUCGAGAUGAUGGGCCUAGCAAACACAGAGCGACAUGUCAACUAUAACCACCCAUCGAUCGGG ..............(((((((.................(((....)))((((((....((.........))((....))...........))))))))))))) (-15.78 = -17.28 + 1.50)

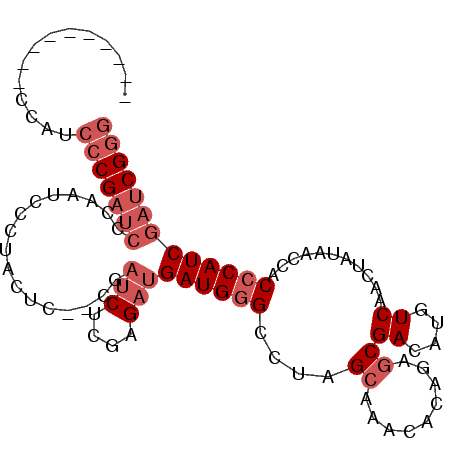
| Location | 15,468,950 – 15,469,041 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -27.20 |
| Energy contribution | -28.29 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
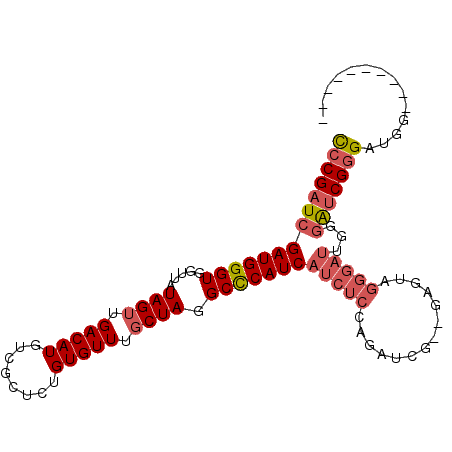
>2L_DroMel_CAF1 15468950 91 - 22407834 CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCGAGAUCG--GAGUAGGGAUAGGGAUCGGGAUGG---------- ((((((((((((((.....((((.(((((........))))).)))).)))))))(((((.......--.....)))))...)))))))....---------- ( -34.90) >DroGri_CAF1 224554 73 - 1 U-CGAUCGAUGGGUGGUUAUAGUUGACAUGUCGUUUUGUGUUUCCUAUGCUCAUCAUCGUCAGG-------------------GACGCGCUGG---------- .-((((.(((((((....((((..((((((......))))))..))))))))))))))).(((.-------------------(...).))).---------- ( -20.50) >DroSec_CAF1 206987 93 - 1 CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCCAGAUUGUAGGGUAGGGAUUGGGAUCGGGAUGG---------- ((((((((((((((.....((((.(((((........))))).)))).)))))))...(((..((......))..)))....)))))))....---------- ( -34.30) >DroSim_CAF1 213078 91 - 1 CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCCAGAUCG--GAGUAGGGAUUGGGAUCGGGAUGG---------- ((((((((((((((.....((((.(((((........))))).)))).)))))))..((((.....)--)))..........)))))))....---------- ( -36.20) >DroEre_CAF1 217645 95 - 1 CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCGAGAUCA--GAGUGGGGAUCGGGAUCGGGAUGG------GGAU (((.((((((((((.....((((.(((((........))))).)))).)))))))(((((((..((.--.....))..)))))))...))).)------)).. ( -37.00) >DroYak_CAF1 215163 101 - 1 CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCGGGAUCG--GAGUGGGGAUUGGGAUCGGGAUGGGGAUGGGGAU (((.((((((((((.....((((.(((((........))))).)))).)))))))(((((((..(((--(........))))..)))))))...))).))).. ( -36.80) >consensus CCCGAUCGAUGGGUGGUUAUAGUUGACAUGUCGCUCUGUGUUUGCUAGGCCCAUCAUCUCCAGAUCG__GAGUAGGGAUUGGGAUCGGGAUGG__________ ((((((((((((((.....((((.(((((........))))).)))).)))))))(((((..............)))))...))))))).............. (-27.20 = -28.29 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:01 2006