| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,465,045 – 15,465,147 |
| Length | 102 |
| Max. P | 0.610309 |

| Location | 15,465,045 – 15,465,147 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
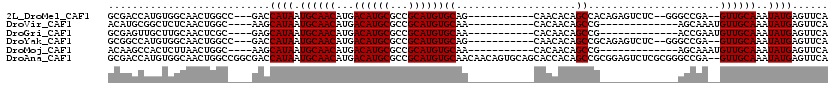
>2L_DroMel_CAF1 15465045 102 - 22407834 GCGACCAUGUGGCAACUGGCC---GACCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAG-----------CAACACAGCCACAGAGUCUC--GGGCCGA--GUUGCAAAUAUGAGUUCA (((..((((((((...(((..---..)))....(((........)))))))))))))).(-----------((((.(.(((...(.....)--.))).).--)))))............. ( -31.90) >DroVir_CAF1 238346 92 - 1 ACAUGCGGCUCUCAACUGGC----AAGCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAA-----------CACAACAGCCG-------------AGCAAAUGUUGCAAAUAUGAGUUCA (((((((((.((((....((----(.......)))....)))...).)))))))))((((-----------((.....((..-------------.))...))))))............. ( -25.10) >DroGri_CAF1 220335 92 - 1 GCGAGUUGCUUGCAACUCGC----GAGCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAA-----------CACAACAGCCG-------------ACCGAAUGUUGCAAAUAUGAGUUCA ((((((((....))))))))----(((((((.((((((((.((((((...))))))((..-----------.......))..-------------.....))))))))..)))..)))). ( -31.60) >DroYak_CAF1 211041 102 - 1 GCGGCCAUGUGGCAACUGGCC---GACCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAG-----------CAACACAGCCGCAGAGUCUC--GGGCCGA--GUUGCAAAUAUGAGUUCA .((((((.((....)))))))---)((((((.((((((..(((.((((..((.((((...-----------...))))))))))..)))((--(...)))--))))))..)))).))... ( -34.80) >DroMoj_CAF1 248742 92 - 1 ACAAGCCACUCUUAACUGGC----AAGCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAA-----------CACAACAGCCG-------------AGCAAAUGUUGCAAAUAUGAGUUCA ....((((........))))----...((((.((((((((....(((.(.((.(((....-----------....))))).)-------------.))).))))))))..))))...... ( -24.50) >DroAna_CAF1 217146 118 - 1 GCGACCAUGUGGCAACUGGCCGGCGACCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAACAACAGUGCAGCACCACAGCCGCGGAGUCUCGCGGGCCGA--GUUGCAAAUAUGAGUUCA ..((((((((.((((((((((.((((.......(((........))).((((.((..(.......)..))((......)).))))....)))).)))).)--)))))..))))).))).. ( -43.50) >consensus GCGACCAUCUGGCAACUGGC____GACCAUAAUGCAACAUGACAUGCGCCGCAUGUGCAA___________CACAACAGCCG_____________ACCGA__GUUGCAAAUAUGAGUUCA ...........................((((.((((((...((((((...))))))((....................))......................))))))..))))...... (-14.00 = -14.00 + 0.00)

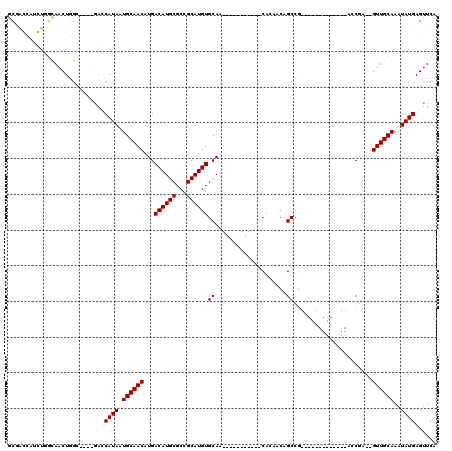
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:58 2006