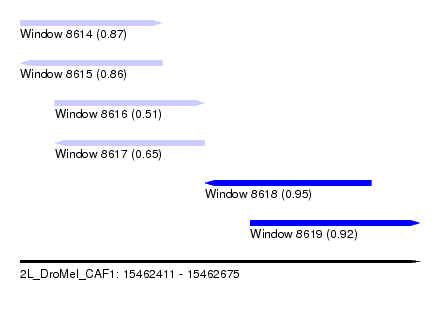
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,462,411 – 15,462,675 |
| Length | 264 |
| Max. P | 0.952056 |

| Location | 15,462,411 – 15,462,505 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.60 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
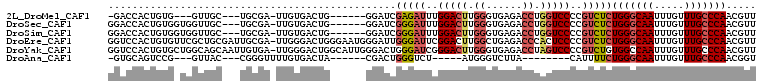
>2L_DroMel_CAF1 15462411 94 + 22407834 -GACCACUGUG---GUUGC---UGCGA-UUGUGACUG------GGAUCGAGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUU -((((.....(---((..(---.....-..)..))).------)).))(((((..(((((.((......)).)))))..)))))(((((((.....)))))))..... ( -34.40) >DroSec_CAF1 200448 98 + 1 GGACCACUGUGGUGGUUGC---UGCGA-UUGUGACUG------GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUU .((((((....))))))..---.(((.-..(.(((.(------((((((((.(((.(.(....)).)))))))))))).))).)(((((((.....))))))).))). ( -40.60) >DroSim_CAF1 206478 98 + 1 GGACCACUGUGGUGGUUGC---UGCGA-UUGUGACUG------GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUU .((((((....))))))..---.(((.-..(.(((.(------((((((((.(((.(.(....)).)))))))))))).))).)(((((((.....))))))).))). ( -40.60) >DroEre_CAF1 211065 107 + 1 GGUCCACUGUGUUCGCUGCGAUUGCGA-UUGGGACUGGGAAUGGGAUUGGGAUUCGGACUUGGCUGAGACCCACUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUU (((((.......((((.......))))-...)))))((((..((((.((((.(((((......))))).)))).))))..))))(((((((.....)))))))..... ( -44.80) >DroYak_CAF1 208340 107 + 1 GGUCCACUGUGCUGGCAGCAAUUGUGA-UUGGGACUGGCAUUGGGACUGGGAUCGGGACUUGGGUGAGACCUAGUCCCCGUCUGUGGCCAAUUUGUUUGCCCAACGUU (((((...((((..((..((((....)-))).).)..))))..))))).((((.((((((.((......)).)))))).)))).(((.(((.....))).)))..... ( -39.80) >DroAna_CAF1 209959 82 + 1 -GUGCAGUCCG---GUUAC---CGGGUUUUGUGACUA------CGACUGGGUCU-----AUGGGUCUUA--------CAUUUUCUGGGCAAUUUGUUUGCCCAACGGU -(((.((.((.---((.((---(.((((.((....))------.)))).)))..-----)).)).))))--------)......(((((((.....)))))))..... ( -25.20) >consensus GGACCACUGUG_UGGUUGC___UGCGA_UUGUGACUG______GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUU ................................................(((((..(((((.((......)).)))))..)))))(((((((.....)))))))..... (-19.54 = -20.60 + 1.06)

| Location | 15,462,411 – 15,462,505 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15462411 94 - 22407834 AACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCUCGAUCC------CAGUCACAA-UCGCA---GCAAC---CACAGUGGUC- .....(((((((.....)))))))((((...((((..((......))..))))...))))(((..------..)))...(-((((.---.....---....))))).- ( -25.20) >DroSec_CAF1 200448 98 - 1 AACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCGAUCC------CAGUCACAA-UCGCA---GCAACCACCACAGUGGUCC ....((((((((.....)))))))).(((.((((.(.((..................)).).)))------).)))....-.....---...(((((....))))).. ( -27.27) >DroSim_CAF1 206478 98 - 1 AACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCGAUCC------CAGUCACAA-UCGCA---GCAACCACCACAGUGGUCC ....((((((((.....)))))))).(((.((((.(.((..................)).).)))------).)))....-.....---...(((((....))))).. ( -27.27) >DroEre_CAF1 211065 107 - 1 AACGUUGGGCAAACAAAUUGCCCAGAGACGGGGAGUGGGUCUCAGCCAAGUCCGAAUCCCAAUCCCAUUCCCAGUCCCAA-UCGCAAUCGCAGCGAACACAGUGGACC ....((((((((.....)))))))).(((.(((((((((........................))))))))).)))(((.-((((.......))))......)))... ( -35.46) >DroYak_CAF1 208340 107 - 1 AACGUUGGGCAAACAAAUUGGCCACAGACGGGGACUAGGUCUCACCCAAGUCCCGAUCCCAGUCCCAAUGCCAGUCCCAA-UCACAAUUGCUGCCAGCACAGUGGACC ..((((.(((..........)))...))))((((((.((......)).)))))).......(((((..(((..((..(((-(....))))..))..)))..).)))). ( -31.50) >DroAna_CAF1 209959 82 - 1 ACCGUUGGGCAAACAAAUUGCCCAGAAAAUG--------UAAGACCCAU-----AGACCCAGUCG------UAGUCACAAAACCCG---GUAAC---CGGACUGCAC- ....((((((((.....))))))))....((--------((.(((....-----.(((...))).------..))).......(((---(...)---)))..)))).- ( -20.50) >consensus AACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCAAUCC______CAGUCACAA_UCGCA___GCAACCA_CACAGUGGACC ..((((((((((.....)))))))...)))((((...((.((......)).))...))))................................................ (-14.62 = -15.48 + 0.86)

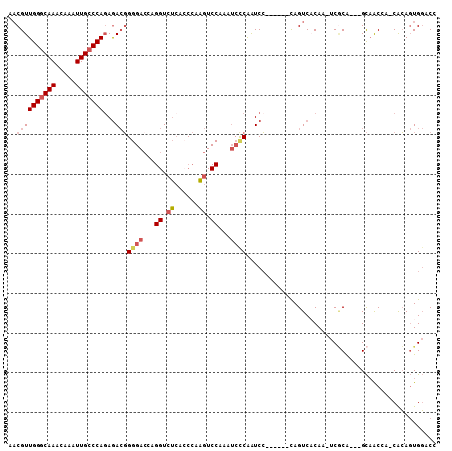
| Location | 15,462,434 – 15,462,533 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -22.51 |
| Energy contribution | -23.73 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15462434 99 + 22407834 UGACUG------GGAUCGAGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCU--- (.((((------(....(((((..(((((.((......)).)))))..)))))(((((((.....)))))))........))))).)..................--- ( -35.20) >DroSec_CAF1 200475 102 + 1 UGACUG------GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU (.((((------(....(((((..(((((.((......)).)))))..)))))(((((((.....)))))))........))))).)........(((......))). ( -35.90) >DroSim_CAF1 206505 102 + 1 UGACUG------GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU (.((((------(....(((((..(((((.((......)).)))))..)))))(((((((.....)))))))........))))).)........(((......))). ( -35.90) >DroEre_CAF1 211095 108 + 1 GGACUGGGAAUGGGAUUGGGAUUCGGACUUGGCUGAGACCCACUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU ..(((((((..((((.((((.(((((......))))).)))).))))..))..(((((((.....)))))))........)))))..........(((......))). ( -38.90) >DroYak_CAF1 208370 108 + 1 GGACUGGCAUUGGGACUGGGAUCGGGACUUGGGUGAGACCUAGUCCCCGUCUGUGGCCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU (((...(((...((((.(((((..((.(((....))).))..))))).)))).(((((((..((((.....)))).)))))))...........)))....))).... ( -37.10) >DroAna_CAF1 209983 89 + 1 UGACUA------CGACUGGGUCU-----AUGGGUCUUA--------CAUUUUCUGGGCAAUUUGUUUGCCCAACGGUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU (.(((.------(((((((..(.-----...)..)...--------.......(((((((.....))))))).))))))...))).)........(((......))). ( -22.40) >consensus UGACUG______GGAUCGGGAUUUGGACUUGGGUGAGACCUGGUCCCCGUCUCUGGGCAAUUUGUUUGCCCAACGUUUGGCCAGUGAAUUUAUUUGCAUUAUCCUGCU ............(((..(((((..(((((.((......)).)))))..)))))(((((((.....))))))).......((.((((....)))).))....))).... (-22.51 = -23.73 + 1.22)

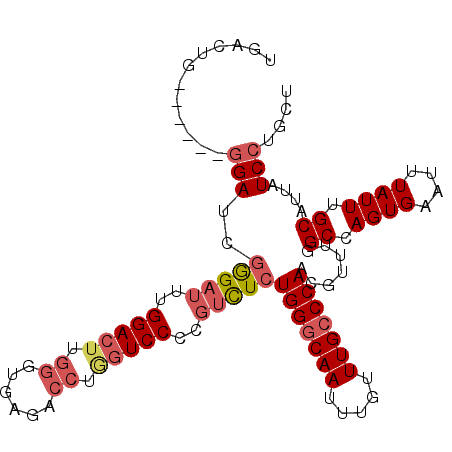
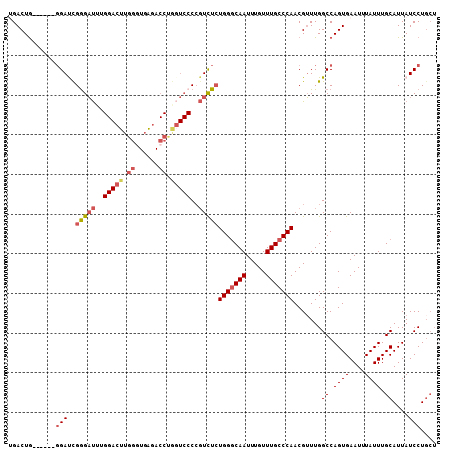
| Location | 15,462,434 – 15,462,533 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -18.96 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15462434 99 - 22407834 ---AGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCUCGAUCC------CAGUCA ---....................(((((........(((((((.....)))))))((((...((((..((......))..))))...))))....)------)))).. ( -28.80) >DroSec_CAF1 200475 102 - 1 AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCGAUCC------CAGUCA .(((......)))..........(((((.....((((((((((.....)))))))...)))((((...((.((......)).))...))))....)------)))).. ( -29.70) >DroSim_CAF1 206505 102 - 1 AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCGAUCC------CAGUCA .(((......)))..........(((((.....((((((((((.....)))))))...)))((((...((.((......)).))...))))....)------)))).. ( -29.70) >DroEre_CAF1 211095 108 - 1 AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGCCCAGAGACGGGGAGUGGGUCUCAGCCAAGUCCGAAUCCCAAUCCCAUUCCCAGUCC .(((......)))..........(((((.....((((((((((.....)))))))...)))((((.((((..((..........))..)))).))))....))))).. ( -34.90) >DroYak_CAF1 208370 108 - 1 AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGGCCACAGACGGGGACUAGGUCUCACCCAAGUCCCGAUCCCAGUCCCAAUGCCAGUCC .(((......)))..........((((((....((((.(((..........)))...))))((((((.((..((...........))..))))))))...)))))).. ( -33.70) >DroAna_CAF1 209983 89 - 1 AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAACCGUUGGGCAAACAAAUUGCCCAGAAAAUG--------UAAGACCCAU-----AGACCCAGUCG------UAGUCA .(((......)))..........(((((.......((((((((.....))))))))...(((--------.......)))-----....)))))..------...... ( -18.50) >consensus AGCAGGAUAAUGCAAAUAAAUUCACUGGCCAAACGUUGGGCAAACAAAUUGCCCAGAGACGGGGACCAGGUCUCACCCAAGUCCAAAUCCCAAUCC______CAGUCA .(((......)))..........((((......((((((((((.....)))))))...)))((((...((.((......)).))...))))...........)))).. (-18.96 = -20.02 + 1.06)

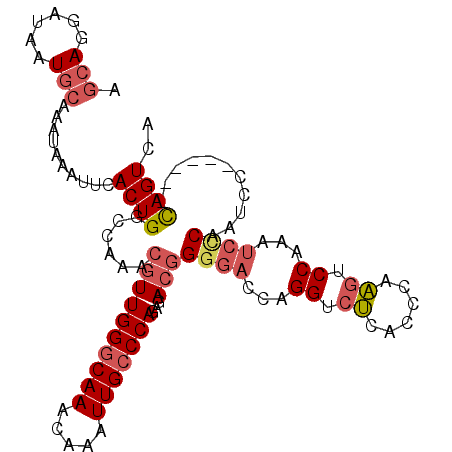
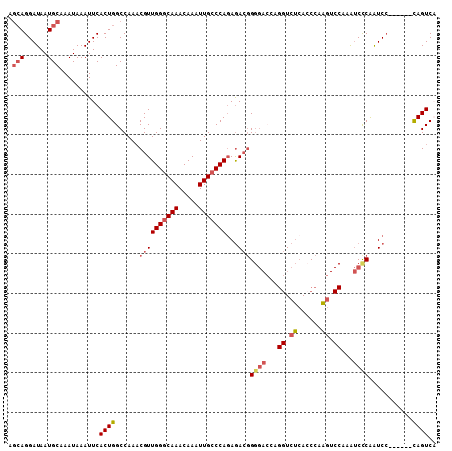
| Location | 15,462,533 – 15,462,643 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -33.83 |
| Energy contribution | -34.58 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15462533 110 - 22407834 CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGCUUUUCGGAUAGUGGCCAACAUUUUAGCCACAGUCAUAGGAACGUGCGACGCCAGCCGGAGC .......((((((.((((...(((........(((((((((...)))))))))..(((.(((((..........))))).)))...)))..))))))))))......... ( -40.20) >DroSec_CAF1 200577 110 - 1 CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGCUUUUCGGAUAGUGGCCACCAUUUUAGCCACAGUCAUUGGAACGUGCGACGCCAGCCGGAGC .......((((((.((((...(((((......(((((((((...)))))))))..(((.(((((..........))))).))).)))))..))))))))))......... ( -41.90) >DroSim_CAF1 206607 110 - 1 CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGCUUUUCGGAUAGUGGCCACCAUUUUAGCCACAGUCAUUGGAACGUGCGACGCCAGCCGGAGC .......((((((.((((...(((((......(((((((((...)))))))))..(((.(((((..........))))).))).)))))..))))))))))......... ( -41.90) >DroEre_CAF1 211203 105 - 1 CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGACUUUUCGGAUAGUGGCCACCAUUUUAGCCGCAGUCAUAGGAACGUGCGACGCCAGCC----- .......((((((.((((...(((..((........))..(((.((((....))))...(((((..........))))).)))...)))..))))))))))....----- ( -33.30) >DroYak_CAF1 208478 105 - 1 CAGCAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGCUUUUCGGAUAGUGGCCACGAUUUUAGCCACAGUCAUAGGAACGUGCGACUCCAGCC----- .......((.(((.((((...(((........(((((((((...)))))))))..(((.(((((..........))))).)))...)))..))))))).))....----- ( -36.60) >DroAna_CAF1 210072 97 - 1 CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGUUUGUGGCA-GGCGGCAGCCG----------ACCGAAAGGAACGUGCAACAUCGGCC--AAC .......((((((.((((...(((..........(((((((...))))))).(((..-(((....))).----------.)))...)))..))))))))))....--... ( -33.80) >consensus CAACAAAGGUGUUUGCACAUGUCCAAGCAAAUAAAAGCCGGGCGCCGGCUUUUCGGAUAGUGGCCACCAUUUUAGCCACAGUCAUAGGAACGUGCGACGCCAGCC__AGC .......((((((.((((...(((........(((((((((...)))))))))..(((.(((((..........))))).)))...)))..))))))))))......... (-33.83 = -34.58 + 0.75)

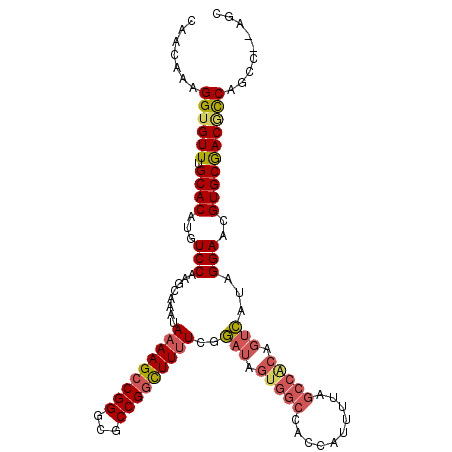
| Location | 15,462,563 – 15,462,675 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -26.39 |
| Energy contribution | -27.92 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15462563 112 + 22407834 UGUGGCUAAAAUGUUGGCCACUAUCCGAAAAGCCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAGCAUUUUUUUAUAUUUUC .((((((((....))))))))......(((((((((...)))))))))(((((((.......((((((.((....))))))))..))))))).................... ( -34.10) >DroSec_CAF1 200607 110 + 1 UGUGGCUAAAAUGGUGGCCACUAUCCGAAAAGCCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAGCAUUUUUUU--AUUUUC .(((((((......)))))))......(((((((((...)))))))))(((((((.......((((((.((....))))))))..)))))))............--...... ( -33.40) >DroSim_CAF1 206637 110 + 1 UGUGGCUAAAAUGGUGGCCACUAUCCGAAAAGCCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAGCAUUUUUUU--AUUUUC .(((((((......)))))))......(((((((((...)))))))))(((((((.......((((((.((....))))))))..)))))))............--...... ( -33.40) >DroEre_CAF1 211228 110 + 1 UGCGGCUAAAAUGGUGGCCACUAUCCGAAAAGUCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAACAUUUUUCU--AUUUUC ...(((((......))))).......((((((((((...))))).((((((((((.......((((((.((....))))))))..))))))))))...))))).--...... ( -28.60) >DroYak_CAF1 208503 110 + 1 UGUGGCUAAAAUCGUGGCCACUAUCCGAAAAGCCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGCUGCAUUUAGCAGAUAAACAUUUUUCU--AUUUUC .(((((((......))))))).....((((((((((...))))).((((((((((.....(((((((((....))))).))))..))))))))))...))))).--...... ( -35.50) >DroAna_CAF1 210100 99 + 1 U----------CGGCUGCCGCC-UGCCACAAACCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAACAUUUUUCU--AUUUUC .----------.....((((..-((((.......))))..)))).((((((((((.......((((((.((....))))))))..)))))))))).........--...... ( -26.40) >consensus UGUGGCUAAAAUGGUGGCCACUAUCCGAAAAGCCGGCGCCCGGCUUUUAUUUGCUUGGACAUGUGCAAACACCUUUGUUGCAUUUAGCAGAUAAACAUUUUUCU__AUUUUC .(((((((......)))))))........(((((((...)))))))(((((((((.......((((((.((....))))))))..))))))))).................. (-26.39 = -27.92 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:56 2006