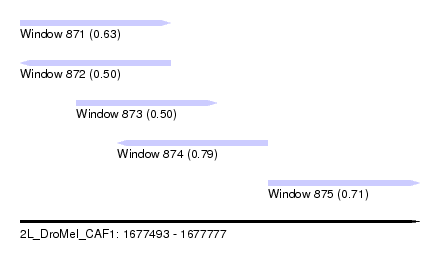
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,677,493 – 1,677,777 |
| Length | 284 |
| Max. P | 0.792406 |

| Location | 1,677,493 – 1,677,600 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677493 107 + 22407834 UCGGCGAAUUCAUCAAUGAUAUAUACAAUAAAAAUACACACGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU---UUUUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAG (((......(((....)))......................(((....)))..---------.))).(((.(((((((---((((((((.....)))))).)-)))))))))))...... ( -24.70) >DroSec_CAF1 3873 105 + 1 UCGGCGAAUUCAUCAAUGAUAUAUACAAUAAAAAUACACACGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU-----UUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAG (((......(((....)))......................(((....)))..---------.))).(((.(((((((-----((((((.....))))))..-.))))))))))...... ( -21.60) >DroSim_CAF1 4863 105 + 1 UCGGCGAAUUCAUCAAUGAUAUAUACAAUAAAAAUACACACGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU-----UUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAG (((......(((....)))......................(((....)))..---------.))).(((.(((((((-----((((((.....))))))..-.))))))))))...... ( -21.60) >DroYak_CAF1 4899 120 + 1 UCGGCGAAUUCAUCAAUGAUAUAUACAAUAAAAAUACACACGCAAUGCUGCAAACGGCGCAAACGGGGGGAUGGUUUUUGUUUUUUCGCCCAGACCGAGACACAAAAACCACCCAAAAAG ((((((...(((....)))(((.....)))..........)))...((((....)))).....))).(((.((((((((((..(((((.......))))).)))))))))))))...... ( -27.40) >consensus UCGGCGAAUUCAUCAAUGAUAUAUACAAUAAAAAUACACACGCAAUGCUGCAA_________ACGGGGGGAUGGUUUU_____UUUCGGCCAGACCGAGACA_AAAAACCACCCAAAAAG (((......(((....)))......................(((....)))............))).(((.(((((((.....((((((.....))))))....))))))))))...... (-21.02 = -21.27 + 0.25)

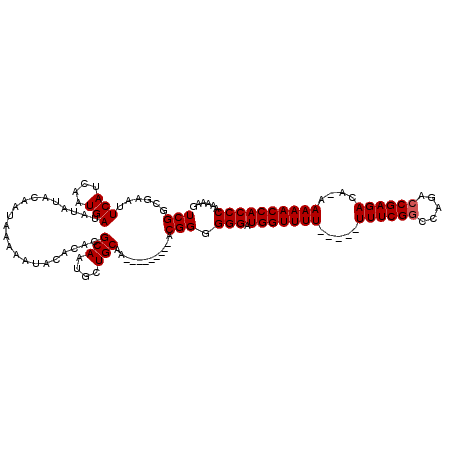
| Location | 1,677,493 – 1,677,600 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677493 107 - 22407834 CUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAAAA---AAAACCAUCCCCCCGU---------UUGCAGCAUUGCGUGUGUAUUUUUAUUGUAUAUAUCAUUGAUGAAUUCGCCGA ......(((((((((((-(...((((.....))))..))---))))))))))...((.---------...((.((.((.((((((((.......)))))))))).)).))....)).... ( -29.30) >DroSec_CAF1 3873 105 - 1 CUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAA-----AAAACCAUCCCCCCGU---------UUGCAGCAUUGCGUGUGUAUUUUUAUUGUAUAUAUCAUUGAUGAAUUCGCCGA ......(((((((((((-(...((((.....))))))-----))))))))))...((.---------...((.((.((.((((((((.......)))))))))).)).))....)).... ( -27.70) >DroSim_CAF1 4863 105 - 1 CUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAA-----AAAACCAUCCCCCCGU---------UUGCAGCAUUGCGUGUGUAUUUUUAUUGUAUAUAUCAUUGAUGAAUUCGCCGA ......(((((((((((-(...((((.....))))))-----))))))))))...((.---------...((.((.((.((((((((.......)))))))))).)).))....)).... ( -27.70) >DroYak_CAF1 4899 120 - 1 CUUUUUGGGUGGUUUUUGUGUCUCGGUCUGGGCGAAAAAACAAAAACCAUCCCCCCGUUUGCGCCGUUUGCAGCAUUGCGUGUGUAUUUUUAUUGUAUAUAUCAUUGAUGAAUUCGCCGA ......(((((((((((((...(((.......)))....))))))))))))).........((.((....((.((.((.((((((((.......)))))))))).)).))....)).)). ( -30.50) >consensus CUUUUUGGGUGGUUUUU_UGUCUCGGUCUGGCCGAAA_____AAAACCAUCCCCCCGU_________UUGCAGCAUUGCGUGUGUAUUUUUAUUGUAUAUAUCAUUGAUGAAUUCGCCGA ......((((((((((......((((.....)))).......))))))))))..................((.((.((.((((((((.......)))))))))).)).)).......... (-23.02 = -23.27 + 0.25)

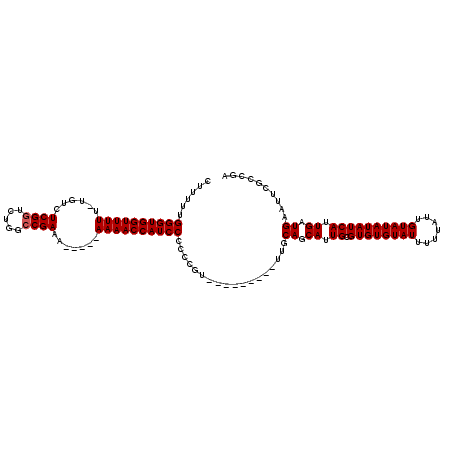
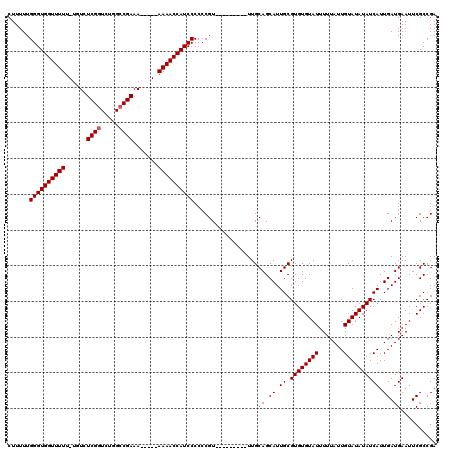
| Location | 1,677,533 – 1,677,633 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -23.43 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677533 100 + 22407834 CGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU---UUUUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAGGAGAAAGAUGUGAAGGCCUAG-------ACCAUUCAUCCU .(((....)))..---------..((((((.(((((((---((((((((.....)))))).)-)))))))))))...............(((((((.....-------.)).)))))))) ( -29.70) >DroSec_CAF1 3913 97 + 1 CGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU-----UUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAGGAGGAAGAAGUGG-GGCCUGG-------ACCAUUCAUCCU .(((....)))..---------.....(((.(((((((-----((((((.....))))))..-.)))))))))).......((((.(((.(((-.......-------.)))))).)))) ( -30.10) >DroSim_CAF1 4903 98 + 1 CGCAAUGCUGCAA---------ACGGGGGGAUGGUUUU-----UUUCGGCCAGACCGAGACA-AAAAACCACCCAAAAAGGAGGAAGAAGUGGAGGCCUGG-------ACCAUUCAUCCU .(((....)))..---------.....(((.(((((((-----((((((.....))))))..-.)))))))))).......((((.(((.(((........-------.)))))).)))) ( -28.00) >DroYak_CAF1 4939 120 + 1 CGCAAUGCUGCAAACGGCGCAAACGGGGGGAUGGUUUUUGUUUUUUCGCCCAGACCGAGACACAAAAACCACCCAAAAAGCUGGAAAAAGUGGAGGCCUGGACCAUGGACCAUUCAUCCU .((...((((....)))).........(((.((((((((((..(((((.......))))).))))))))))))).....)).(((...(((((...((........)).)))))..))). ( -35.00) >consensus CGCAAUGCUGCAA_________ACGGGGGGAUGGUUUU_____UUUCGGCCAGACCGAGACA_AAAAACCACCCAAAAAGGAGGAAGAAGUGGAGGCCUGG_______ACCAUUCAUCCU .(((....))).............((((((.(((((((.....((((((.....))))))....))))))))))...............(((((((.............)).)))))))) (-23.43 = -23.74 + 0.31)

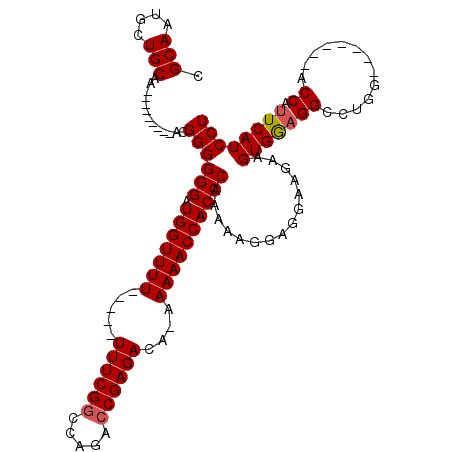
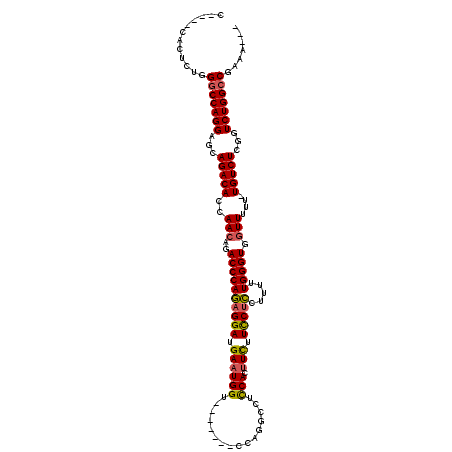
| Location | 1,677,562 – 1,677,669 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -30.73 |
| Energy contribution | -30.79 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677562 107 - 22407834 C----CACUCUGGGCCAGGAGCAGACACCAACAGACCCAGAGGAUGAAUGGU-------CUAGGCCUUCACAUCUUUCUCCUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAAAA- .----.......(((((((...(((((..(((..((((((((((((((.(((-------....)))))))........)))))..))))).)))...-)))))...)))))))......- ( -37.80) >DroSec_CAF1 3942 104 - 1 C----CACUCUGGGCCAGGAGCAGACACCAAAAGACCCAGAGGAUGAAUGGU-------CCAGGCC-CCACUUCUUCCUCCUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAA--- .----.......(((((((...(((((..(((..((((((((((.((((((.-------.......-))).))).))))).....)))))..)))..-)))))...)))))))....--- ( -39.20) >DroSim_CAF1 4932 105 - 1 C----CACUCUGGGCCAGGAGCAGACACCAACAGACCCAGAGGAUGAAUGGU-------CCAGGCCUCCACUUCUUCCUCCUUUUUGGGUGGUUUUU-UGUCUCGGUCUGGCCGAAA--- .----.......(((((((...(((((..(((..((((((((((.(((.(((-------....))).....))).))))).....))))).)))...-)))))...)))))))....--- ( -38.40) >DroYak_CAF1 4979 120 - 1 CACAGCACUCUGGGCCAGGAGCAGACACCAACAGACCCACAGGAUGAAUGGUCCAUGGUCCAGGCCUCCACUUUUUCCAGCUUUUUGGGUGGUUUUUGUGUCUCGGUCUGGGCGAAAAAA ....(((((.(((((((((........))..((..((....)).))..))))))).)))(((((((..(((.....(((.((....)).))).....)))....)))))))))....... ( -37.10) >consensus C____CACUCUGGGCCAGGAGCAGACACCAACAGACCCAGAGGAUGAAUGGU_______CCAGGCCUCCACUUCUUCCUCCUUUUUGGGUGGUUUUU_UGUCUCGGUCUGGCCGAAA___ ............(((((((...(((((..(((..((((((((((.((((((................))).))).))))).....))))).)))....)))))...)))))))....... (-30.73 = -30.79 + 0.06)

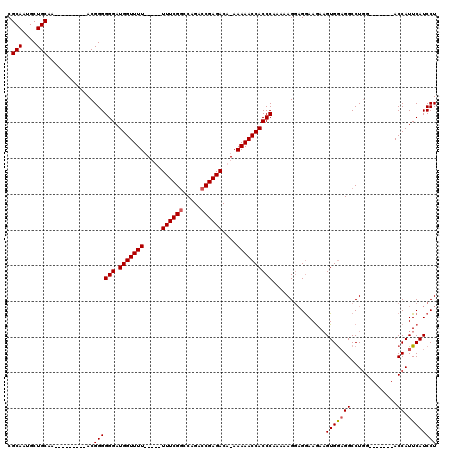
| Location | 1,677,669 – 1,677,777 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -32.07 |
| Energy contribution | -32.01 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677669 108 + 22407834 UUGCAAAAGGGGCGGAUGGGGGAGUGACUGGACCAUAGUUAUCGGCCUGGACCAGUGACAUUUCUGGCCAGGCAUUCAGCUCGUUCGG------------CUCCUGUGUGUUACUUCUAU ..(((..(((((((((((((.(((((((((.....)))))....((((((.((((........))))))))))))))..))))))).)------------)))))...)))......... ( -43.80) >DroSec_CAF1 4046 108 + 1 UUGCAAAAGGGGCGGAAGGGGGAGUGACUGGGCCAUAGUUAUCGGCCUGGACCAGUGACAUUUCUGGCCAGGCAUUCAGCUCGUUCGG------------CUCCUGUGUGUUACUUCUAU ..(((..((((((.(((.(((((.((((((.....)))))))).((((((.((((........))))))))))......))).))).)------------)))))...)))......... ( -44.70) >DroSim_CAF1 5037 108 + 1 UUGCAAAAGGGGCGGUAGGGGGAGUGACUGGGCCAUAGUUAUCGGCCUGGACCAGUGACAUUUCUGGCCAGGCAUUCAGCUCGUUCGG------------CUCCUGUGUGUUACUUCUAU ..(((..((((((.(..(((.(((((((((.....)))))....((((((.((((........))))))))))))))..)))...).)------------)))))...)))......... ( -40.50) >DroYak_CAF1 5099 118 + 1 UGGC--AGGGGGCGGAGGGGGGAGUGGCUGGGCCAUAGUUAUCGUCCUGGACCAGUGACAUUUCUGCCCAGGCAUUCAGCUCGUUCAGCUCUGGCUCCAGCUCCUGUGUGUUACUUCUAU ..((--((((((((((.(((.(((((.((((((...((..((.((((((...))).)))))..)))))))).)))))..))).))).)).(((....))))))))))............. ( -45.80) >consensus UUGCAAAAGGGGCGGAAGGGGGAGUGACUGGGCCAUAGUUAUCGGCCUGGACCAGUGACAUUUCUGGCCAGGCAUUCAGCUCGUUCGG____________CUCCUGUGUGUUACUUCUAU .......(((((..(..(.(((((...((((((...((((....((((((.((((........))))))))))....)))).))))))............))))).)...)..))))).. (-32.07 = -32.01 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:52 2006