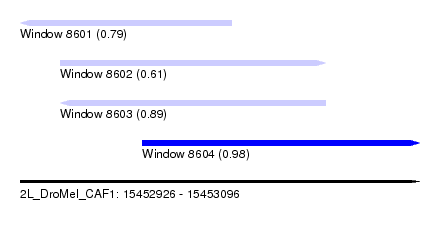
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,452,926 – 15,453,096 |
| Length | 170 |
| Max. P | 0.977832 |

| Location | 15,452,926 – 15,453,016 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
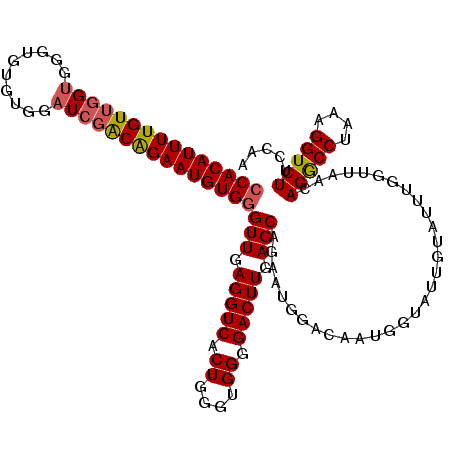
>2L_DroMel_CAF1 15452926 90 - 22407834 ACAUUCUGUCGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGCCAACCUUGGGGUCCCAUCAUCAUCGAUCGAUUC ....((.(((((((((((...............))))))))((((((((.((.....)).)).))))))............))).))... ( -25.66) >DroSec_CAF1 191052 90 - 1 ACAUUCUGUGGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCAUCAUCAUCGAUCGAUUC ....((.(((((((((((...............))))))))((((((((.(((...))).)).))))))........))).))....... ( -27.06) >DroSim_CAF1 197006 90 - 1 ACAUUCUGUCGAACCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCAUCAUCAUCGAUCGAUUC ....((.(((((.(((((...............)))))...((((((((.(((...))).)).))))))..........))))).))... ( -25.36) >DroEre_CAF1 201310 90 - 1 ACAUUCCGCCGAUCCACACACCCACCAACAAAAUGUGGACCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCAUCAUCAUCGAUCGAUUC .........(((((.......((((.........))))...((((((((.(((...))).)).))))))............))))).... ( -24.30) >DroYak_CAF1 198580 90 - 1 ACAUUCUGUCUAUCCCCACACCCACCAACAAAAUGUGUAUCGCCCCAGGAUGCGUGCCAAGCUUGGGGUCCCAUCAUCAUCGAUCGAUUG .((.((.(((.......((((.............))))...((((((((..(....)....))))))))............))).)).)) ( -16.42) >consensus ACAUUCUGUCGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCAUCAUCAUCGAUCGAUUC .......(((((((.......((((.........))))...((((((((.((.....))..))))))))............))))))).. (-18.84 = -19.84 + 1.00)



| Location | 15,452,943 – 15,453,056 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -36.64 |
| Energy contribution | -37.16 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15452943 113 + 22407834 UGGGACCCCAAGGUUGGCACGCAUCCUGGGGCGAUCCACAUUUUGUUGGUGGGUGUGUGGAUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACA .....(((((.((.((.....)).)))))))(((((((((((((((((((..........)))))))))))))))))))..(((.(((...(....).....)))....))). ( -41.50) >DroSec_CAF1 191069 113 + 1 UGGGACCCCAAGGUUGCCACGCAUCCUGGGGCGAUCCACAUUUUGUUGGUGGGUGUGUGGAUCCACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACA ..((((((((.((.(((...))).)))))))...))).((((((((((.(.(((((.(.(((((((.....))))))).).).)))).).)(....)...))))))))).... ( -41.20) >DroSim_CAF1 197023 113 + 1 UGGGACCCCAAGGUUGCCACGCAUCCUGGGGCGAUCCACAUUUUGUUGGUGGGUGUGUGGUUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACA (((....)))(((((.((((.((.(((....((((((((((((((((((............)))))))))))))))))))))....)).)))).))))).............. ( -42.20) >DroEre_CAF1 201327 112 + 1 UGGGACCCCAAGGUUGCCACGCAUCCUGGGGCGGUCCACAUUUUGUUGGUGGGUGUGUGGAUCGGCGGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUCAGCAGAAUGGAC- ...(.(((((.((.(((...))).))))))))...(((((((((((((((..........)))))))))))))))(((((((((.((....)).))))))))).........- ( -44.90) >DroYak_CAF1 198597 112 + 1 UGGGACCCCAAGCUUGGCACGCAUCCUGGGGCGAUACACAUUUUGUUGGUGGGUGUGGGGAUAGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGAC- (((....))).(((..(..(.(((((..((.(((..(((((((((((.((..........)).)))))))))))..)))...))...))))).)...)..))).........- ( -33.10) >consensus UGGGACCCCAAGGUUGCCACGCAUCCUGGGGCGAUCCACAUUUUGUUGGUGGGUGUGUGGAUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACA .....(((((.((.((.....)).)))))))(((((((((((((((((((..........)))))))))))))))))))..(((.(((...(....).....)))....))). (-36.64 = -37.16 + 0.52)

| Location | 15,452,943 – 15,453,056 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -24.08 |
| Energy contribution | -25.88 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15452943 113 - 22407834 UGUCCAUUCUGCUCAAGUCCCCACCCAGUGACCUCAACCCACAUUCUGUCGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGCCAACCUUGGGGUCCCA ...........................(.((((((((....(((((((.(((((((((...............)))))))))...))))))).((....)).)))))))).). ( -30.06) >DroSec_CAF1 191069 113 - 1 UGUCCAUUCUGCUCAAGUCCCCACCCAGUGACCUCAACCCACAUUCUGUGGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCA ...........................(.(((((((((((.(((((((.(((((((((...............)))))))).)..))))))).).)).....)))))))).). ( -30.86) >DroSim_CAF1 197023 113 - 1 UGUCCAUUCUGCUCAAGUCCCCACCCAGUGACCUCAACCCACAUUCUGUCGAACCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCA ...........................(.(((((((((((.(((((((.(((.(((((...............))))).)))...))))))).).)).....)))))))).). ( -28.66) >DroEre_CAF1 201327 112 - 1 -GUCCAUUCUGCUGAAGUCCCCACCCAGUGACCUCAACCCACAUUCCGCCGAUCCACACACCCACCAACAAAAUGUGGACCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCA -.........((((...........))))((((((((.((((..(((..((.((((((...............)))))).)).....)))...)))).....))))))))... ( -27.56) >DroYak_CAF1 198597 112 - 1 -GUCCAUUCUGCUCAAGUCCCCACCCAGUGACCUCAACCCACAUUCUGUCUAUCCCCACACCCACCAACAAAAUGUGUAUCGCCCCAGGAUGCGUGCCAAGCUUGGGGUCCCA -..........................(.((((((((..(.(((((((........((((.............))))........))))))).).((...)))))))))).). ( -18.51) >consensus UGUCCAUUCUGCUCAAGUCCCCACCCAGUGACCUCAACCCACAUUCUGUCGAUCCACACACCCACCAACAAAAUGUGGAUCGCCCCAGGAUGCGUGGCAACCUUGGGGUCCCA ...........................(.(((((((((((.(((((((.(((((((((...............)))))))))...))))))).).)).....)))))))).). (-24.08 = -25.88 + 1.80)


| Location | 15,452,978 – 15,453,096 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15452978 118 + 22407834 CCACAUUUUGUUGGUGGGUGUGUGGAUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACAAUGGUAUUGUAUUUGGUUAACAGGCCUAAAGGUCUUCCAA (((((((((((((((..........)))))))))))))))(((.(((((.((....)).))))).)))....(((((((...........)))......(((((....))))))))). ( -37.00) >DroSec_CAF1 191104 118 + 1 CCACAUUUUGUUGGUGGGUGUGUGGAUCCACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACAAUGGUAUUGUAUUUGGUUAACAGGCCUAAAGGUCUUUCAA (((((((((((....((((......)))))))))))))))(((.(((((.((....)).))))).))).(((.((((...(((.((((.........))))))).....))))))).. ( -34.60) >DroSim_CAF1 197058 118 + 1 CCACAUUUUGUUGGUGGGUGUGUGGUUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACAAUGGUAUUGUAUUUGGUUAACAGGCCUAAAGGUCUUCCAA ((((((((((((((............))))))))))))))(((.(((((.((....)).))))).)))....(((((((...........)))......(((((....))))))))). ( -35.80) >DroEre_CAF1 201362 103 + 1 CCACAUUUUGUUGGUGGGUGUGUGGAUCGGCGGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUCAGCAGAAUGGAC-------------UUGGUUAAGAGACCUAAAGGUCUA--CU (((((((((((((((..........)))))))))))))))(((((((((.((....)).)))))))))....(((((-------------((((((....))))...)))))))--.. ( -39.80) >DroYak_CAF1 198632 105 + 1 ACACAUUUUGUUGGUGGGUGUGGGGAUAGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGAC-------------UUGGUUAAGAGACCUAAAGGUCUUCGCC .(((((((((((.((..........)).))))))))))).(((.(((((.((....)).))))).))).........-------------..(((...((((((....)))))).))) ( -30.00) >consensus CCACAUUUUGUUGGUGGGUGUGUGGAUCGACAGAAUGUGGGUUGAGGUCACUGGGUGGGGACUUGAGCAGAAUGGACAAUGGUAUUGUAUUUGGUUAACAGGCCUAAAGGUCUUCCAA (((((((((((((((..........)))))))))))))))(((.(((((.((....)).))))).)))...............................(((((....)))))..... (-30.62 = -30.86 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:42 2006