
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,452,761 – 15,452,854 |
| Length | 93 |
| Max. P | 0.986321 |

| Location | 15,452,761 – 15,452,854 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
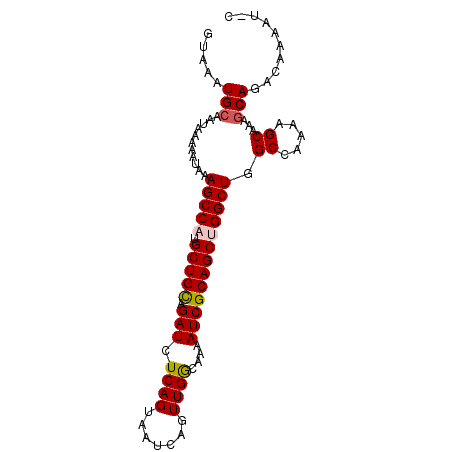
>2L_DroMel_CAF1 15452761 93 + 22407834 GCAUUUUGUCUGCUUUGCUUUUGGCCAGCCAGCUGCGAUUUUGCCAACUGAUUACUGAGAUCUACAGCAUGGCUUUAUUUUUAUUGCAUUUAC (((........(((........))).(((((((((.((((((..............))))))..)))).)))))..........)))...... ( -19.14) >DroSec_CAF1 190887 93 + 1 GUAUUUUGUCUGCUUUGCUGUUGGCCAGCCGGCUGCGAUUUUUUCAACUGAUUACUGAGAUCUGCAGCAUGGCUUUAUUUUUAUUGCAUUUAC (((....(((.((......)).))).((((((((((((((((.((....)).....)))))).))))).)))))..........)))...... ( -23.00) >DroSim_CAF1 196841 93 + 1 GCAUUUUGUCUGCUUUGCUUUUGGCCAGCCGGCUGCGAUUUUGCCAACUGAUUACUGAGAUCUGCAGCAUGGCUUUAUUUUUAUUGCAUUUAC (((........(((........))).((((((((((((((((..............)))))).))))).)))))..........)))...... ( -22.94) >DroEre_CAF1 201155 83 + 1 ----------UGGUUCGCUUUUGGCCAGCCAGCUGCGAUUUUGCCAACUGAUUACUGAGAUCUGCAGCAUGGCUUUAUUUUUAUUGCAUUUAC ----------(((((.......)))))(((((((((((((((..............)))))).))))).)))).................... ( -21.44) >DroYak_CAF1 198425 83 + 1 ----------UGCUUUGCUUUUGGCCAGCCAGCUGCGAUUUUGCCAACUGAUUACUGAGAUCUGCAGCAUGGCUUUAUUUUUAUUGCAUUUAC ----------(((...((.....)).((((((((((((((((..............)))))).))))).)))))...........)))..... ( -21.84) >consensus G_AUUUUGUCUGCUUUGCUUUUGGCCAGCCAGCUGCGAUUUUGCCAACUGAUUACUGAGAUCUGCAGCAUGGCUUUAUUUUUAUUGCAUUUAC ..........(((...((.....)).((((((((((((((((..............)))))).))))).)))))...........)))..... (-19.36 = -19.52 + 0.16)



| Location | 15,452,761 – 15,452,854 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15452761 93 - 22407834 GUAAAUGCAAUAAAAAUAAAGCCAUGCUGUAGAUCUCAGUAAUCAGUUGGCAAAAUCGCAGCUGGCUGGCCAAAAGCAAAGCAGACAAAAUGC ((...(((...........(((((.(((((.(((.((((.......))))....))))))))))))).((.....))...))).))....... ( -20.20) >DroSec_CAF1 190887 93 - 1 GUAAAUGCAAUAAAAAUAAAGCCAUGCUGCAGAUCUCAGUAAUCAGUUGAAAAAAUCGCAGCCGGCUGGCCAACAGCAAAGCAGACAAAAUAC ((...(((...........((((..(((((.(((.((((.......))))....)))))))).)))).((.....))...))).))....... ( -20.40) >DroSim_CAF1 196841 93 - 1 GUAAAUGCAAUAAAAAUAAAGCCAUGCUGCAGAUCUCAGUAAUCAGUUGGCAAAAUCGCAGCCGGCUGGCCAAAAGCAAAGCAGACAAAAUGC ((...(((................(((((.......))))).(((((((((.........)))))))))...........))).))....... ( -19.90) >DroEre_CAF1 201155 83 - 1 GUAAAUGCAAUAAAAAUAAAGCCAUGCUGCAGAUCUCAGUAAUCAGUUGGCAAAAUCGCAGCUGGCUGGCCAAAAGCGAACCA---------- .....(((...........(((((.(((((.(((.((((.......))))....)))))))))))))........))).....---------- ( -17.41) >DroYak_CAF1 198425 83 - 1 GUAAAUGCAAUAAAAAUAAAGCCAUGCUGCAGAUCUCAGUAAUCAGUUGGCAAAAUCGCAGCUGGCUGGCCAAAAGCAAAGCA---------- .....(((...........(((((.(((((.(((.((((.......))))....))))))))))))).((.....))...)))---------- ( -19.90) >consensus GUAAAUGCAAUAAAAAUAAAGCCAUGCUGCAGAUCUCAGUAAUCAGUUGGCAAAAUCGCAGCUGGCUGGCCAAAAGCAAAGCAGACAAAAU_C .....(((...........(((((.(((((.(((.((((.......))))....))))))))))))).((.....))...))).......... (-17.06 = -17.34 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:39 2006