
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,449,721 – 15,449,864 |
| Length | 143 |
| Max. P | 0.981945 |

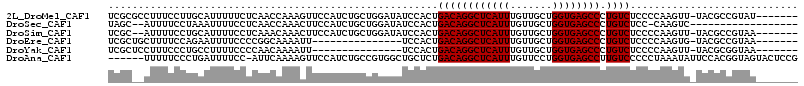
| Location | 15,449,721 – 15,449,828 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.24 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15449721 107 + 22407834 UCGCGCCUUUCCUUGCAUUUUUCUCAACCAAAGUUCCAUCUGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCCGUAU------- .((.((.....((((............(((.((......))..))).........((((((((((((.......)))))).))))))....))))..-...))))...------- ( -24.10) >DroSec_CAF1 187935 94 + 1 UAGC--AUUUUCCUAAAUUUUCCUCAACCAAACUUCCAUCUGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCC-CAAGUC------------------ ....--............................((((.....))))........((((((((((((.......)))))).))))))...-......------------------ ( -19.10) >DroSim_CAF1 193854 105 + 1 UCGC--AUUUUCCUGCAUUUUCCUCAAACAAACUUCCAUCUGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCCGUAA------- ..((--(......))).............((((((........(((....)))..((((((((((((.......)))))).)))))).....)))))-).........------- ( -23.50) >DroEre_CAF1 198251 92 + 1 UCGCUGCUUUUCCAGAAUUUUCCCCGGCAAAAUU---------------UCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUG-UACGCCGUAA------- ...(((......))).........((((...(((---------------(.....((((((((((((.......)))))).)))))).....)))).-...))))...------- ( -24.00) >DroYak_CAF1 195460 92 + 1 UCGCUCCUUUCCCUGCCUUUUCCCCAACAAAAUU---------------UCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCGGUAA------- .............((((.................---------------......((((((((((((.......)))))).)))))).......((.-...)))))).------- ( -19.30) >DroAna_CAF1 197311 108 + 1 ------UUUUUCCCUGAUUUUCC-AUUCAAAAGUUCCAUCUGCCGUGGCUGCUCUGACAGGCUCAUUUGUUCCUGGUGAGCCUUGUCCCCCUAAAUAUUCCACGGUAGUACUCCG ------........(((......-..)))..(((.....(((((((((.......((((((((((((.......)))))))).))))............))))))))).)))... ( -29.61) >consensus UCGC__CUUUUCCUGCAUUUUCCUCAACAAAACUUCCAUCUGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU_UACGCCGUAA_______ .......................................................((((((((((((.......)))))))).))))............................ (-16.23 = -16.23 + -0.00)

| Location | 15,449,761 – 15,449,864 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -20.53 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15449761 103 + 22407834 UGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCCGUAU--------CUUCCAAAAUCCGGGCCUCAUUAGCCCGCCCAAGGC ...(((....)))..((((((((((((.......)))))).))))))..........-...((((...--------...).......(((((.......))))).....))) ( -28.00) >DroSec_CAF1 187973 90 + 1 UGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCC-CAAGUC---------------------UCCAAAAUCCGGGCCUCAUUAGCCCGCCUAAGGC .((((((.((.....((((((((((((.......)))))).))))))...-...)).---------------------)))......(((((.......))))).....))) ( -28.70) >DroSim_CAF1 193892 103 + 1 UGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCCGUAA--------CUUCCAAAAUCCGGGCCUCAUUAGCCCGCCUAAGGC .(((((((.......((((((((((((.......)))))).)))))).....(((((-........))--------))).....))))((((.......))))......))) ( -30.40) >DroEre_CAF1 198285 94 + 1 ---------UCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUG-UACGCCGUAA--------CUUUCACAAUCCGGGCCUCAUUAGCCCGCCUAAGGC ---------..((((((((((((((((.......)))))).))))))......))))-...(((((..--------.....))....(((((.......))))).....))) ( -28.60) >DroYak_CAF1 195494 94 + 1 ---------UCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU-UACGCGGUAA--------CUUCCACAAUCCGAGCCUUAUUAGCCCGUCUAAGGC ---------......((((((((((((.......)))))).))))))..........-...(.((...--------...)).).......((((((..........)))))) ( -23.10) >DroAna_CAF1 197344 111 + 1 UGCCGUGGCUGCUCUGACAGGCUCAUUUGUUCCUGGUGAGCCUUGUCCCCCUAAAUAUUCCACGGUAGUACUCCGAAAUCCGAAAUCCGGCCCUCAUUAGCCAACCC-GGGC ((((((((.......((((((((((((.......)))))))).))))............)))))))).......((...(((.....)))...))....(((.....-.))) ( -32.71) >consensus UGCUGGAUAUCCACUGACAGGCUCAUUUGUUGCUGGUGAGCCCUGUCUCCCCAAGUU_UACGCCGUAA________CUUCCAAAAUCCGGGCCUCAUUAGCCCGCCUAAGGC ..........((...((((((((((((.......)))))))).))))........................................(((((.......))))).....)). (-20.53 = -21.37 + 0.83)


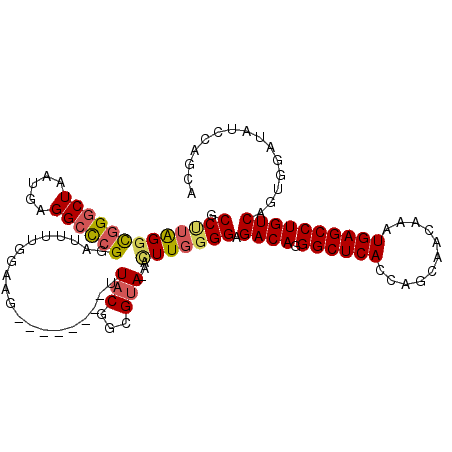
| Location | 15,449,761 – 15,449,864 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15449761 103 - 22407834 GCCUUGGGCGGGCUAAUGAGGCCCGGAUUUUGGAAG--------AUACGGCGUA-AACUUGGGGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGAUAUCCAGCA .(((..((((((((.....)))))).((.(((....--------...))).)).-..))..))).((((((.((((...........))))))))))..(((....)))... ( -33.70) >DroSec_CAF1 187973 90 - 1 GCCUUAGGCGGGCUAAUGAGGCCCGGAUUUUGGA---------------------GACUUG-GGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGAUAUCCAGCA (((...)))(((((.....)))))((((.(((..---------------------..)...-...((((((.((((...........))))))))))....)).)))).... ( -31.00) >DroSim_CAF1 193892 103 - 1 GCCUUAGGCGGGCUAAUGAGGCCCGGAUUUUGGAAG--------UUACGGCGUA-AACUUGGGGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGAUAUCCAGCA .(((((((((((((.....))))))..........(--------(....))...-..))))))).((((((.((((...........))))))))))..(((....)))... ( -33.60) >DroEre_CAF1 198285 94 - 1 GCCUUAGGCGGGCUAAUGAGGCCCGGAUUGUGAAAG--------UUACGGCGUA-CACUUGGGGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGA--------- .(((((((((((((.....))))))...((((...(--------(....)).))-))))))))).((((((.((((...........))))))))))......--------- ( -33.60) >DroYak_CAF1 195494 94 - 1 GCCUUAGACGGGCUAAUAAGGCUCGGAUUGUGGAAG--------UUACCGCGUA-AACUUGGGGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGA--------- .((((((.((((((.....))))))....((((...--------...))))...-...)))))).((((((.((((...........))))))))))......--------- ( -31.10) >DroAna_CAF1 197344 111 - 1 GCCC-GGGUUGGCUAAUGAGGGCCGGAUUUCGGAUUUCGGAGUACUACCGUGGAAUAUUUAGGGGGACAAGGCUCACCAGGAACAAAUGAGCCUGUCAGAGCAGCCACGGCA (((.-((.((((((......))))))((((((.....))))))....))((((.........(....)...((((..((((..(....)..))))...))))..))))))). ( -35.90) >consensus GCCUUAGGCGGGCUAAUGAGGCCCGGAUUUUGGAAG________UUACGGCGUA_AACUUGGGGAGACAGGGCUCACCAGCAACAAAUGAGCCUGUCAGUGGAUAUCCAGCA .(((((((((((((.....))))))....................(((...)))...))))))).((((.((((((...........))))))))))............... (-23.39 = -23.80 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:36 2006