| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,437,151 – 15,437,321 |
| Length | 170 |
| Max. P | 0.667552 |

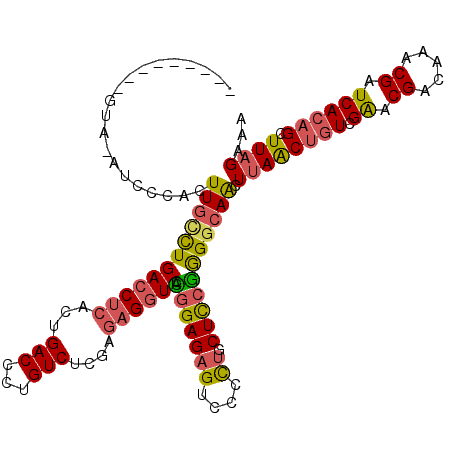
| Location | 15,437,151 – 15,437,256 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -15.61 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15437151 105 + 22407834 UGCAAAUUCGAUCAAAUAAUGUACAAACAUUAUUGUGGAUUAUCCC-AGUGGUGUUUCU--------------UUUUAAGUUUCUAAGCUGUGAUCGUUUGUCGUUCGACAGUUAAGUUG ........((((((..........(((((((((((.((.....)))-))))))))))..--------------.....((((....)))).)))))).(((((....)))))........ ( -23.70) >DroPse_CAF1 181346 91 + 1 CUCGAACUCGAUCAGAAAAUGCCCAAACAUUAUGCUGGAUUAUCCC-AGUGGUGUUU----------------UUCGGACUUUCUUGGCUGUGAUCGUUUGUCGUUUG------------ ..(((...(((((((((((((((..........(((((......))-))))))))))----------------))(((.((.....)))))))))))....)))....------------ ( -22.80) >DroEre_CAF1 189628 105 + 1 UGCAAAUUCGAUCAAAUAAUGUACAAACAUUAUUGUGGAUUAUCCC-AGUGGUGUUUCU--------------UUUUAGGUUUCUAAGCUGUGAUCGUUUGUCGUUCGACAGCUAAACUG .((.....((((((..........(((((((((((.((.....)))-))))))))))..--------------.(((((....)))))...))))))..((((....))))))....... ( -25.50) >DroYak_CAF1 190354 105 + 1 UGCAAAUUCGAUCAAAUAAUGUUCAAACAUUAUGGUGGAUUAUCCU-AGUGGUGUUUCU--------------UUUUAAGUUUCAAAGCUGUGAUCGUUUGUCGUCCGACAGUUAAACUG ........((((((..........(((((((((...((.....)).-.)))))))))..--------------.....((((....)))).)))))).(((((....)))))........ ( -20.30) >DroAna_CAF1 191516 120 + 1 CGCAAAUUCGAUCAAAUAAUGUACAAACAUUAUGCUGGAUUAUCCUGAGUGGUGUUUCUCUUUUUUUUUUUUUUUUUGGUUUUCUAAACUCUGAUCGUUUGUCGUUCAACUGUUAAACUG ((((((..((((((..........(((((((((...((.....))...))))))))).................(((((....)))))...)))))))))).))................ ( -19.60) >DroPer_CAF1 182409 90 + 1 CUCGAACUCGAUCAGAAAAUGCCCAAACAUUAUGCUGGAUUAUCCC-AGUGGUGUUU----------------UUCGGACUCUCUUGGCUCUGAUCGUUUGUCGU-UG------------ ..(((...((((((((....(((.(((((((..(((((......))-))))))))))----------------...((.....)).)))))))))))....))).-..------------ ( -26.30) >consensus CGCAAAUUCGAUCAAAUAAUGUACAAACAUUAUGCUGGAUUAUCCC_AGUGGUGUUUCU______________UUUUAAGUUUCUAAGCUGUGAUCGUUUGUCGUUCGACAGUUAAACUG .(((((..((((((..........(((((((((...((.....))...))))))))).................((((......))))...))))))))))).................. (-15.61 = -14.70 + -0.91)

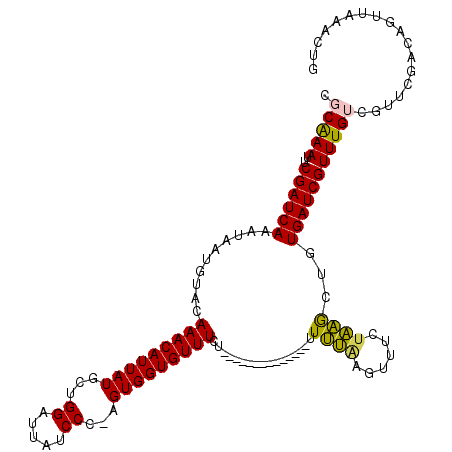

| Location | 15,437,151 – 15,437,256 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15437151 105 - 22407834 CAACUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAACUUAAAA--------------AGAAACACCACU-GGGAUAAUCCACAAUAAUGUUUGUACAUUAUUUGAUCGAAUUUGCA .........((((....))))..((((((..................--------------............-.((.....))..((((((((....))))))))))))))........ ( -17.30) >DroPse_CAF1 181346 91 - 1 ------------CAAACGACAAACGAUCACAGCCAAGAAAGUCCGAA----------------AAACACCACU-GGGAUAAUCCAGCAUAAUGUUUGGGCAUUUUCUGAUCGAGUUCGAG ------------....(((....((((((.......(((((((((((----------------........((-((......)))).......)))))))..))))))))))...))).. ( -20.37) >DroEre_CAF1 189628 105 - 1 CAGUUUAGCUGUCGAACGACAAACGAUCACAGCUUAGAAACCUAAAA--------------AGAAACACCACU-GGGAUAAUCCACAAUAAUGUUUGUACAUUAUUUGAUCGAAUUUGCA .......((((((....))))..(((((((((.((((....))))..--------------.(......).))-)((.....))..((((((((....)))))))))))))).....)). ( -19.70) >DroYak_CAF1 190354 105 - 1 CAGUUUAACUGUCGGACGACAAACGAUCACAGCUUUGAAACUUAAAA--------------AGAAACACCACU-AGGAUAAUCCACCAUAAUGUUUGAACAUUAUUUGAUCGAAUUUGCA .........((((....))))..((((((..................--------------............-.((.....))...(((((((....))))))).))))))........ ( -16.10) >DroAna_CAF1 191516 120 - 1 CAGUUUAACAGUUGAACGACAAACGAUCAGAGUUUAGAAAACCAAAAAAAAAAAAAAAAAGAGAAACACCACUCAGGAUAAUCCAGCAUAAUGUUUGUACAUUAUUUGAUCGAAUUUGCG ..(((((.....)))))(.(((((((((((((((..........................(((........))).((.....))))).((((((....))))))))))))))..))))). ( -19.60) >DroPer_CAF1 182409 90 - 1 ------------CA-ACGACAAACGAUCAGAGCCAAGAGAGUCCGAA----------------AAACACCACU-GGGAUAAUCCAGCAUAAUGUUUGGGCAUUUUCUGAUCGAGUUCGAG ------------..-.(((....((((((((.....(((.(((((((----------------........((-((......)))).......))))))).)))))))))))...))).. ( -24.86) >consensus CAGUUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAACUCAAAA______________AGAAACACCACU_GGGAUAAUCCACCAUAAUGUUUGUACAUUAUUUGAUCGAAUUUGCA .......................((((((..............................................((.....))...(((((((....))))))).))))))........ (-12.07 = -12.18 + 0.11)

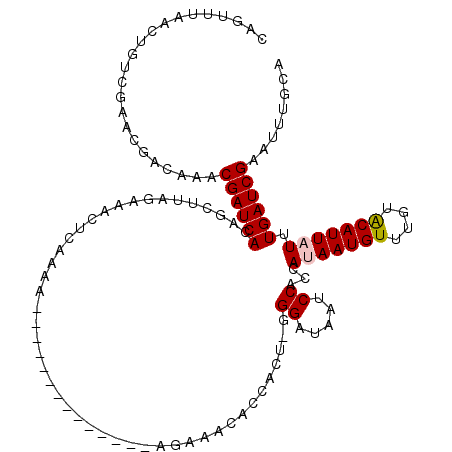

| Location | 15,437,216 – 15,437,321 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15437216 105 - 22407834 GUAGCA------------ACUUGCCUGACCUCACUGACCCUGUCUCGAGAGGUCACGGAGAGUCCCCUGCUCGGGGGCAACUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAA ((.((.------------(((..(((((((((...(((...)))....))))))).))..)))((((.....)))))).))((((((((.((.((.....)).)))))).))))... ( -32.90) >DroSec_CAF1 182891 117 - 1 GUAGCACUGGUAGAUCCCACUUGACUGACCUCUCUGACCCUGUCUCGAGAGGUCACGGAGAGUCCCCUGCUCCGGGGCAACUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAA ((.((.((((.((.....((((..((((((((((.(((...)))..))))))))).)..))))......)))))).)).))((((((((.((.((.....)).)))))).))))... ( -36.60) >DroSim_CAF1 188794 117 - 1 GUAGCACUGGUAGAUCCCACUUGCCUGACCUCACUGACCCUGUCUCGAGAGGUCACGGAGAGUCCCCUGCUCCGGGGCAACUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAA ..(((....((.((((....((((((((((((...(((...)))....)))))).(((((((....)).)))))))))))......((((....))))...)))))).)))...... ( -36.50) >DroEre_CAF1 189693 109 - 1 --------CGUAUAUCCCACUUGUCUGACCUCACUGACCCUGUCUCGAGAGGUCACGGAGAGUCCUCUGCUCCGAGGCAGUUUAGCUGUCGAACGACAAACGAUCACAGCUUAGAAA --------............((((((((((((...(((...)))....)))))).(((((((....)).)))))))))))(((((((((.((.((.....)).))))))).)))).. ( -33.70) >DroYak_CAF1 190419 109 - 1 --------CGUUAAUGGCACUUUCCUGACCUCAGAGACCCUGUCUCGAGAGGUCAUGGAGAGUCCUCUGCUCCGGGGCAGUUUAACUGUCGGACGACAAACGAUCACAGCUUUGAAA --------.(((((.((.((((((((((((((.(((((...)))))..))))))).))))))))).(((((....))))).)))))((((....))))................... ( -42.70) >DroAna_CAF1 191596 102 - 1 -------------CUCCCACUUGCUUGACCUCAAUGACCCUGUCCCGA--GUUCAUGGAGAGACCCUACCUUCAGGGCAGUUUAACAGUUGAACGACAAACGAUCAGAGUUUAGAAA -------------((((((...(((((....((.......))...)))--))...))).))).((((......))))...(((((...((((.((.....)).))))...))))).. ( -18.50) >consensus _________GUA_AUCCCACUUGCCUGACCUCACUGACCCUGUCUCGAGAGGUCACGGAGAGUCCCCUGCUCCGGGGCAACUUAACUGUCGAACGACAAACGAUCACAGCUUAGAAA ....................((((((((((((...(((...)))....)))))).(((((((....)).))))))))))).((((((((.((.((.....)).)))))).))))... (-23.03 = -23.15 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:33 2006