
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
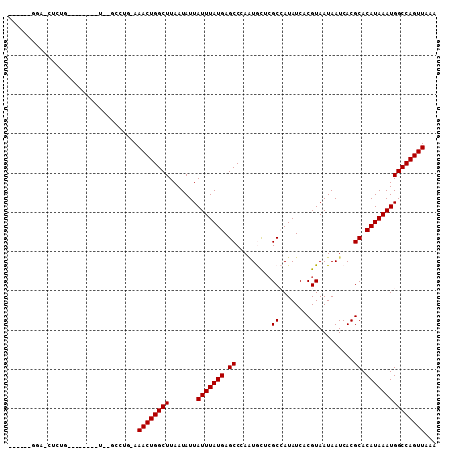
| Location | 15,435,861 – 15,435,959 |
| Length | 98 |
| Max. P | 0.934894 |

| Location | 15,435,861 – 15,435,959 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -15.93 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
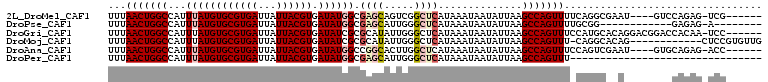
>2L_DroMel_CAF1 15435861 98 + 22407834 ------CGA-CUCUGGAC----AUUCGCCUGAAAACUGGCUUAAUAUUAUUUAUGAGCCGACUGCUCGCCAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA ------...-.((.((.(----....))).)).((((((((.......((((((((((.....)))).........(((.......)))...))))))))))))))... ( -18.11) >DroPse_CAF1 180150 88 + 1 --------U-CUCUC------------CCGCAAAACUGGCUUAAUAUUAUUUAUGAGCCCAAUGCUCGCCAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA --------.-.....------------......((((((((.......((((((((((.....)))).........(((.......)))...))))))))))))))... ( -16.91) >DroGri_CAF1 193690 102 + 1 ------GGA-UUGUGGUCCGUCCUGUGCAUGGAAACUGGCUUAAUAUUAUUUAUGAGCCCAAUAUGCGCGAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA ------(((-(....)))).(((.......)))((((((((.......(((((((.((......((((.(....).))))........)).)))))))))))))))... ( -25.75) >DroMoj_CAF1 224501 96 + 1 CAACACGGAG------------CUGUGCCUG-AAACUGGCUUAAUAUUAUUUAUGAGCCCAAUAUGCGCGAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA ........((------------(((.(((..-.....((((((.((.....)))))))).....((((.(((...........))).)))).......))))))))... ( -22.70) >DroAna_CAF1 190401 98 + 1 ------GGU-CUCUGCAC----AUUCGACUGGAAACUGGCUUAAUAUUAUUUAUGAGCCAAGUGCCGGCCAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA ------...-....((((----........(....)(((((((.((.....))))))))).)))).((((((....(((.......))).......))))))....... ( -23.90) >DroPer_CAF1 181209 77 + 1 --------------------------------AAACUGGCUUAAUAUUAUUUAUGAGCCCAAUGCUCGCCAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA --------------------------------.((((((((.......((((((((((.....)))).........(((.......)))...))))))))))))))... ( -16.91) >consensus ______GGA_CUCUG________U__GCCUG_AAACUGGCUUAAUAUUAUUUAUGAGCCCAAUGCUCGCCAUAUCACGUAAUAAUCACGCACAUAAAUGGCCAGUUAAA .................................((((((((.......(((((((.((.........((........)).........)).)))))))))))))))... (-15.93 = -15.93 + -0.00)


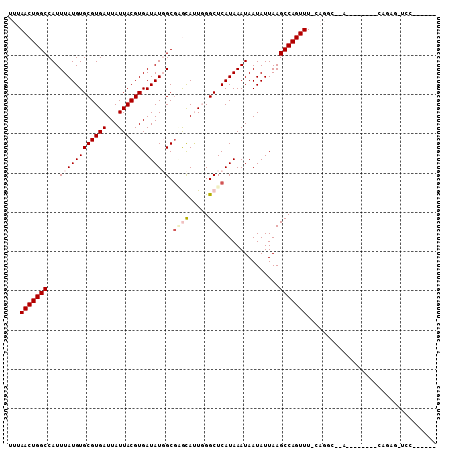
| Location | 15,435,861 – 15,435,959 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
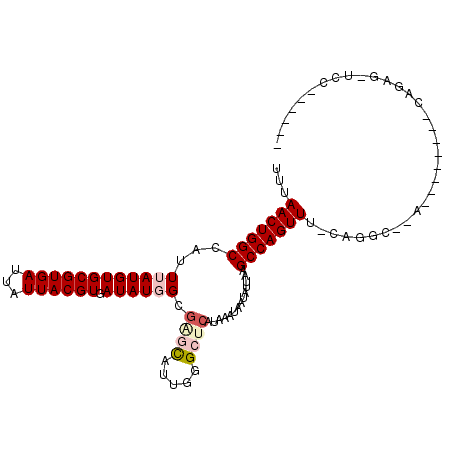
>2L_DroMel_CAF1 15435861 98 - 22407834 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUGGCGAGCAGUCGGCUCAUAAAUAAUAUUAAGCCAGUUUUCAGGCGAAU----GUCCAGAG-UCG------ .....((((.(((((....((((((...))))))((..((((((((.....)))).(((......))).))))....))....))))----).))))..-...------ ( -24.60) >DroPse_CAF1 180150 88 - 1 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUGGCGAGCAUUGGGCUCAUAAAUAAUAUUAAGCCAGUUUUGCGG------------GAGAG-A-------- ...(((((((...((((((((((((...)))))).)))))).((((.....))))..............)))))))......------------.....-.-------- ( -21.50) >DroGri_CAF1 193690 102 - 1 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUCGCGCAUAUUGGGCUCAUAAAUAAUAUUAAGCCAGUUUCCAUGCACAGGACGGACCACAA-UCC------ ......(((((.(((.((((((((.......((((....)))).......((((..............))))......))))))))))).)).)))...-...------ ( -24.94) >DroMoj_CAF1 224501 96 - 1 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUCGCGCAUAUUGGGCUCAUAAAUAAUAUUAAGCCAGUUU-CAGGCACAG------------CUCCGUGUUG .......((((....(((((((((((.(((....))))))))))))))..))))...............(((.....-..))).(((------------(.....)))) ( -26.60) >DroAna_CAF1 190401 98 - 1 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUGGCCGGCACUUGGCUCAUAAAUAAUAUUAAGCCAGUUUCCAGUCGAAU----GUGCAGAG-ACC------ ......(((((((.....(((((((...)))))))..)))))))((((((((((..............)))))).(((.....))).----))))....-...------ ( -24.54) >DroPer_CAF1 181209 77 - 1 UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUGGCGAGCAUUGGGCUCAUAAAUAAUAUUAAGCCAGUUU-------------------------------- ...(((((((...((((((((((((...)))))).)))))).((((.....))))..............))))))).-------------------------------- ( -21.50) >consensus UUUAACUGGCCAUUUAUGUGCGUGAUUAUUACGUGAUAUGGCGAGCAUUGGGCUCAUAAAUAAUAUUAAGCCAGUUU_CAGGC__A________CAGAG_UCC______ ...(((((((...((((((((((((...)))))).)))))).((((.....))))..............)))))))................................. (-16.42 = -17.28 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:30 2006