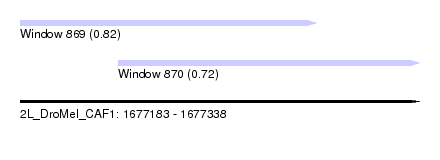
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,677,183 – 1,677,338 |
| Length | 155 |
| Max. P | 0.819740 |

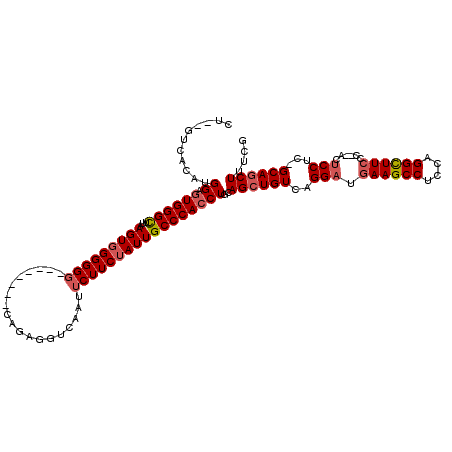
| Location | 1,677,183 – 1,677,298 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -26.89 |
| Energy contribution | -28.01 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677183 115 + 22407834 CU--GUCACAUGGUCUGGGCUUAAGUGGGGGGGUGGGUAGUAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCAGGCUUCCC--ACUCCUC-GCAGCUUUCG ..--.((((..(((....)))...))))(.((((((((((((((((......)))))))))))))))).)((((((.((((.((((((....))))))..--..)))).-)))))).... ( -56.20) >DroSec_CAF1 3572 107 + 1 AU--GUCACAUGGAGUGGGCUUAAGUGGGGGG--------CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCGCCAGGCUUCCC--ACUCCUC-GCAGCUUUCG ..--.((((.....))))......(..(((((--------((((((......))))...))))).))..)((((((.((((.((((((....))))))..--..)))).-)))))).... ( -43.80) >DroSim_CAF1 4562 107 + 1 CU--GUCACAUGGAGUGGGCUUAAGUGGGGGG--------CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCGGGCUUCCC--ACUCCUC-GCAGCUUUCG ((--((.....((((((((((...(..(((((--------((((((......))))...))))).))..)))).........((((((....))))))))--)))))..-))))...... ( -44.70) >DroYak_CAF1 4593 114 + 1 CUCAAUCCAUUGGAGUGGGUUUAAGUGGGGG------UAGUAGAGGUCACUUCUUCAAUUGCCCACCUGCAGCUGUCAGGUUGAAGCCACCCGGUUUACCCAACCCCUCUGCAACUUUCG ...........((((.(((((...(((((((------((((.((((......)))).)))))))((((((....).))))).....))))..((....)).))))))))).......... ( -37.00) >consensus CU__GUCACAUGGAGUGGGCUUAAGUGGGGGG________CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCAGGCUUCCC__ACUCCUC_GCAGCUUUCG ...........((.((((((...(((((((((...................)))))))))))))))))..((((((..(((.((((((....))))))......)))...)))))).... (-26.89 = -28.01 + 1.12)

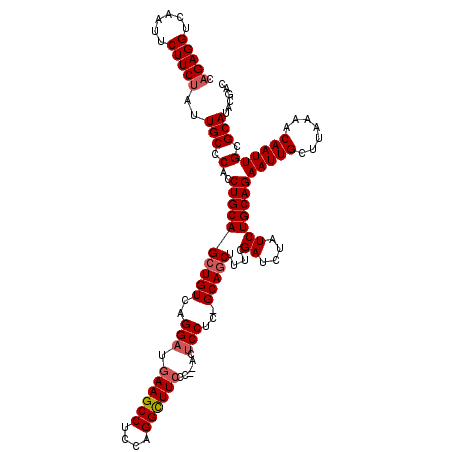
| Location | 1,677,221 – 1,677,338 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -25.06 |
| Energy contribution | -25.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1677221 117 + 22407834 UAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCAGGCUUCCC--ACUCCUC-GCAGCUUUCGAUCUAUCUGCAGAAUUGCUUAAAACAAUUGCGCAUACGAC ((((((......)))))).(((.(..((((((((((.((((.((((((....))))))..--..)))).-)))))....((....)))))))(((((.......)))))).)))...... ( -35.20) >DroSec_CAF1 3602 117 + 1 CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCGCCAGGCUUCCC--ACUCCUC-GCAGCUUUCGAUCUAUCUGCAGAAUUGCUUAAAACAAUUGCGCAUACGAC .(((((......)))))..(((.(..((((((((((.((((.((((((....))))))..--..)))).-)))))....((....)))))))(((((.......)))))).)))...... ( -34.80) >DroSim_CAF1 4592 117 + 1 CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCGGGCUUCCC--ACUCCUC-GCAGCUUUCGAUCUAUCUGCAGAAUUGCUUAAAACAAUUGCGCAUACGAC .(((((......)))))..(((.(..((((((((((.((((.((((((....))))))..--..)))).-)))))....((....)))))))(((((.......)))))).)))...... ( -34.40) >DroYak_CAF1 4627 120 + 1 UAGAGGUCACUUCUUCAAUUGCCCACCUGCAGCUGUCAGGUUGAAGCCACCCGGUUUACCCAACCCCUCUGCAACUUUCGAUCUAUCUGCAGAAUUGCUUAAAACAAUUGCGCAUACGAC ..((((......))))............((((.(((..((((((((((....)))))...)))))..((((((..............))))))..........))).))))......... ( -23.94) >consensus CAGAGGUCAAUUCUUCUAUUGCCCACCUGCAGCUGUCAGGAUGAAGCCUCCAGGCUUCCC__ACUCCUC_GCAGCUUUCGAUCUAUCUGCAGAAUUGCUUAAAACAAUUGCGCAUACGAC .(((((......)))))..(((.(..((((((((((..(((.((((((....))))))......)))...)))))....((....)))))))(((((.......)))))).)))...... (-25.06 = -25.88 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:48 2006