| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,412,685 – 15,412,787 |
| Length | 102 |
| Max. P | 0.540611 |

| Location | 15,412,685 – 15,412,787 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
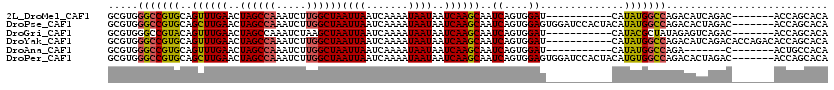
>2L_DroMel_CAF1 15412685 102 - 22407834 GCGUGGGCCGUGCAGUUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAU-----------CAUAUGGCCAGACAUCAGAC-------ACCAGCACA ((((((((((((..((((((..((((((.....))))))((((.......))))..)))))).(((....)))-----------..))))))).(....)...)-------))..))... ( -24.50) >DroPse_CAF1 156937 113 - 1 GCGUGGGCCGUGCAGCUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAGUGGAUCCACUACAUAUGGCCAGACACUAGAC-------ACCAGCACA ((((((((((((..((((((..((((((.....))))))((((.......))))..))))))....((((((.....))))))...)))))))...(....).)-------))..))... ( -34.90) >DroGri_CAF1 166630 102 - 1 GCGUGGGCCGUACAGUUUGAACUAGCCAAAUCUAAGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAU-----------CAUACGCUAUAGAGUCAGAC-------ACCAGCACA ((.(((.(.(.((.((((((..((((.........))))((((.......))))..))))))....((((...-----------....)))).....))).)..-------.)))))... ( -15.40) >DroYak_CAF1 165630 109 - 1 GCGUGGGCCGUGCAGUUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAU-----------CAUAUGGCCAGACAUCAGACACCAGACACCAGCACA ((((((((((((..((((((..((((((.....))))))((((.......))))..)))))).(((....)))-----------..))))))).....((........)))))..))... ( -24.70) >DroAna_CAF1 165800 95 - 1 GCGUGGGCCGUGCAGUUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAU-----------CAUAUGGCCAGA-------C-------ACUGCCACA ((((((((((((..((((((..((((((.....))))))((((.......))))..)))))).(((....)))-----------..)))))))...-------)-------)).)).... ( -24.50) >DroPer_CAF1 157442 113 - 1 GCGUGGGCCGUGCAGCUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAGUGGAUCCACUACAUGUGGCCAGACACUAGAC-------ACCAGCACA ((....)).((((.((((((..((((((.....))))))((((.......))))..)))))).....(((.((((..(((((.....))))..)..))))...)-------))..)))). ( -33.80) >consensus GCGUGGGCCGUGCAGUUUGAACUAGCCAAAUCUUGGCUAAUUAAUCAAAAUAAUAAUCAAGCAAUCAGUGGAU___________CAUAUGGCCAGACAUCAGAC_______ACCAGCACA .....(((((((..((((((..((((((.....))))))((((.......))))..))))))..((....))..............)))))))........................... (-19.38 = -19.27 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:20 2006