| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,402,207 – 15,402,313 |
| Length | 106 |
| Max. P | 0.958620 |

| Location | 15,402,207 – 15,402,313 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
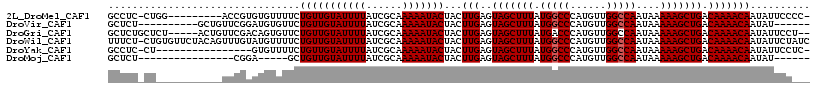
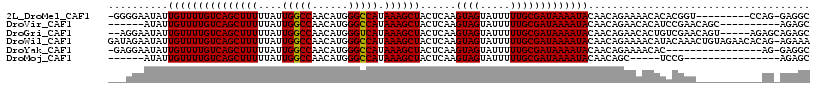
>2L_DroMel_CAF1 15402207 106 + 22407834 GCCUC-CUGG---------ACCGUGUGUUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUCCCC- .....-..((---------(.....((((((..((.(((((((......))))))).))....((((((((.(((((......)))))....)))))).))))))))....)))..- ( -24.10) >DroVir_CAF1 173308 101 + 1 GCUCU----------GCUGUUCGGAUGUGUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAU------ ..(((----------(.....))))..((((.(((((((((((......))))))).........((((((.(((((......)))))....)))))))))).))))....------ ( -22.60) >DroGri_CAF1 155998 110 + 1 GCUCUGCUCU-----ACUGUUCGACAGUGUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGACCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUCCU-- ...(((((((-----((((.....)))))....((.(((((((......))))))).))..))))))............(((..((.........))..))).............-- ( -22.20) >DroWil_CAF1 254134 116 + 1 UUUCU-CUGUGUUCUACAGUUUGUAUGUUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUCUAUC .....-(((((...)))))(((((.....(((.((.(((((((......))))))).))..)))(((((((.(((((......)))))....))))))))))))............. ( -23.80) >DroYak_CAF1 155214 99 + 1 GCCUC-CU----------------GUGUUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUCCUC- .....-..----------------.((((((..((.(((((((......))))))).))....((((((((.(((((......)))))....)))))).)))))))).........- ( -21.30) >DroMoj_CAF1 183289 90 + 1 GCUCU----------------CGGA-----GCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAU------ ((((.----------------..))-----))(((((((((((......)))))))...(((..(((((((.(((((......)))))....))))))).)))))))....------ ( -22.70) >consensus GCUCU_CU___________UUCGGAUGUUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUCC___ ................................(((((((((((......)))))))...(((..(((((((.(((((......)))))....))))))).))))))).......... (-18.02 = -18.18 + 0.17)
| Location | 15,402,207 – 15,402,313 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.25 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
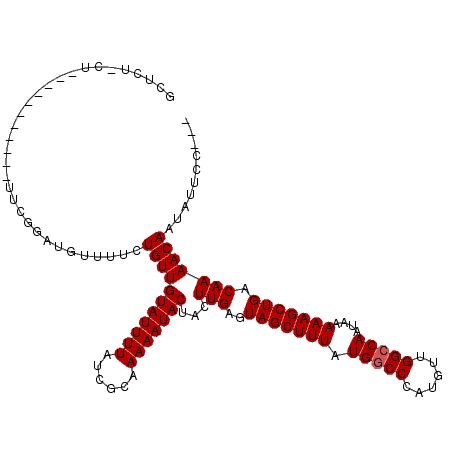
>2L_DroMel_CAF1 15402207 106 - 22407834 -GGGGAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAACACACGGU---------CCAG-GAGGC -..(((....((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..)))))).....)---------))..-..... ( -28.00) >DroVir_CAF1 173308 101 - 1 ------AUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAACACAUCCGAACAGC----------AGAGC ------....((((((((.((((((....(((((......))))).)))))).........(((((((......))))))).))))))))............----------..... ( -26.20) >DroGri_CAF1 155998 110 - 1 --AGGAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGUCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAACACUGUCGAACAGU-----AGAGCAGAGC --.........((((((..((((((....(((((......))))).)))))).(((..((.(((((((......))))))).)).....((((.....))))-----.))))))))) ( -26.90) >DroWil_CAF1 254134 116 - 1 GAUAGAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAACAUACAAACUGUAGAACACAG-AGAAA ((((((((...))))))))((((((....(((((......))))).)))))).(((..((.(((((((......))))))).))........(((.....))).......)-))... ( -28.60) >DroYak_CAF1 155214 99 - 1 -GAGGAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAACAC----------------AG-GAGGC -.........((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..)))))).----------------..-..... ( -24.00) >DroMoj_CAF1 183289 90 - 1 ------AUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGC-----UCCG----------------AGAGC ------....(((((((((((((((....(((((......))))).))))))......((((.....))))))))))))).....((-----((..----------------.)))) ( -26.10) >consensus ___GGAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAACAUACGAA___________AG_AGAGC ..........(((((((((((((((....(((((......))))).))))))......((((.....)))))))))))))..................................... (-23.39 = -23.25 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:16 2006