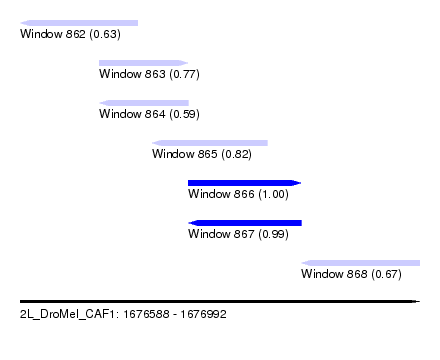
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,676,588 – 1,676,992 |
| Length | 404 |
| Max. P | 0.995306 |

| Location | 1,676,588 – 1,676,707 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.61 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676588 119 - 22407834 CCUUUUUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA-AGCAAACGUUAUGCCAGAAACUAAAUUAAGAUUGAAAUGUAAAACUGAAAGCGUUUUACACAAUUAUUGAACGCUCGAGAC ...((((((((((((...)))))))(.((((..(((((-(((....))).))))).............(((((...((((((((.......))))))))))))).)))).)...))))). ( -32.40) >DroSec_CAF1 3045 116 - 1 CCUU-UUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA-AGCAAACGUUAUGCCAGAAACUAAAUUAAGAUUGAAAUGUAAAACUGAAAGCGUUUUACACAAUUAUUGAACGCUCG--AC ....-...(((((((...))))))).(((....(((((-(((....))).))))).........(((((((((...((((((((.......))))))))))))).))))......)--)) ( -30.90) >DroSim_CAF1 3932 116 - 1 CCUU-UUGGCCGGACUUUGUCCGGCGGUCAGAAUGGGA-AGCAAACGUUAUGCCAGAAACUAAAUUAAGAUUGAAAUGUAAAACUGAAAGCGUUUUACACAAUUAUUGAACGCUCG--AC ((((-((((((((((...)))))))...))))).))((-.((....(((........)))....(((((((((...((((((((.......))))))))))))).))))..)))).--.. ( -26.90) >DroYak_CAF1 4074 117 - 1 CCUU-UUAGACGGACUUUGUCCGGCGGUCAGAAAGGCAGAGCAAACGUUAUGCCAGAAACUAAAUUAAGAUUGAAAUGUAAAACUGAAAGCGUUUUACACAAUUAUUGAACGCUCG--AC ....-.....(((((...)))))(((.((((...((((.(((....))).))))..............(((((...((((((((.......))))))))))))).)))).)))...--.. ( -29.60) >consensus CCUU_UUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA_AGCAAACGUUAUGCCAGAAACUAAAUUAAGAUUGAAAUGUAAAACUGAAAGCGUUUUACACAAUUAUUGAACGCUCG__AC ..........(((((...)))))(((.((((...((((.(((....))).))))..............(((((...((((((((.......))))))))))))).)))).)))....... (-27.67 = -27.92 + 0.25)

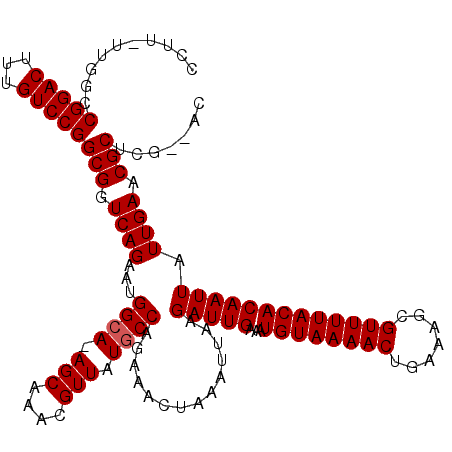
| Location | 1,676,668 – 1,676,758 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676668 90 + 22407834 U-UGCCAUUCUGACCGCCGGACAAAGUCCGGCCAAAAAGGGGUUCCGUCCGAAU----------------------UUGGGUAUUUACGGAUUUAUUGCGAGGGGUAGGCAAA (-((((((((((((((((((((...)))))))(......))))).(((..((((----------------------(((........)))))))...))).))))).))))). ( -29.30) >DroSec_CAF1 3123 85 + 1 U-UGCCAUUCUGACCGCCGGACAAAGUCCGGCCAA-AAGGGGUUCCGUCCGAAAAAAUG--------------------------AAGGGAUUUAUUACGAGGGGUAGCCAAA (-((((.(((.(((.(((((((...)))))))...-..((....))))).)))..((((--------------------------((....))))))......)))))..... ( -23.50) >DroYak_CAF1 4152 112 + 1 UCUGCCUUUCUGACCGCCGGACAAAGUCCGUCUAA-AAGGGGUUUUGUCCGCAAAAAAUUACGAGGGGGAUGGUGCAUGGGUAUUUACGGAUUUGUUUCGAGGGGCAGGCAAA ((((((((((.....((.(((((((..((.(....-.).))..)))))))))..........((((.(((((((((....))))).....)))).)))))))))))))).... ( -36.70) >consensus U_UGCCAUUCUGACCGCCGGACAAAGUCCGGCCAA_AAGGGGUUCCGUCCGAAAAAA____________________UGGGUAUUUACGGAUUUAUUACGAGGGGUAGGCAAA ..((((..((.....(((((((...)))))))......(..((...(((((....................................)))))..))..)))..))))...... (-19.59 = -19.82 + 0.23)

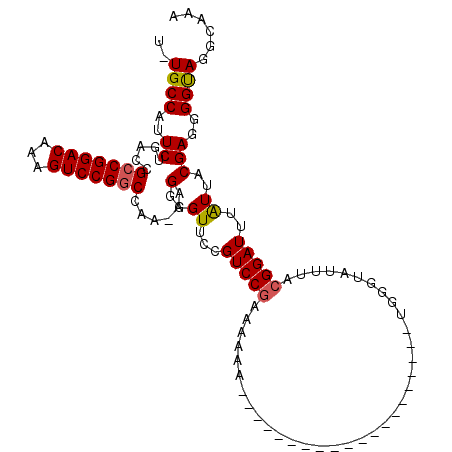
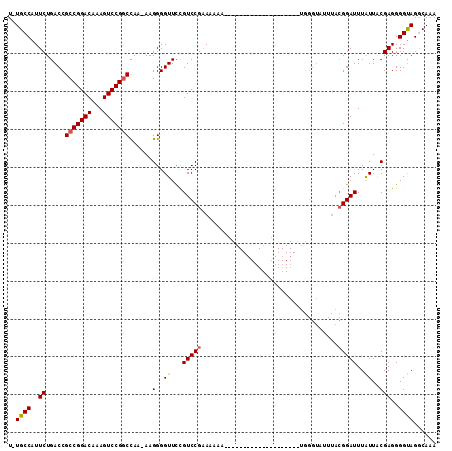
| Location | 1,676,668 – 1,676,758 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676668 90 - 22407834 UUUGCCUACCCCUCGCAAUAAAUCCGUAAAUACCCAA----------------------AUUCGGACGGAACCCCUUUUUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA-A .(((((.....((.((......(((((......((..----------------------....)))))))...........(((((((...))))))).)).))...))))-) ( -25.20) >DroSec_CAF1 3123 85 - 1 UUUGGCUACCCCUCGUAAUAAAUCCCUU--------------------------CAUUUUUUCGGACGGAACCCCUU-UUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA-A (((((((...............(((.((--------------------------(........))).))).......-...(((((((...))))))))))))))......-. ( -23.20) >DroYak_CAF1 4152 112 - 1 UUUGCCUGCCCCUCGAAACAAAUCCGUAAAUACCCAUGCACCAUCCCCCUCGUAAUUUUUUGCGGACAAAACCCCUU-UUAGACGGACUUUGUCCGGCGGUCAGAAAGGCAGA .....(((((..((...........(((........)))......((((..((((....))))(((((((..((...-......))..))))))))).))...))..))))). ( -26.10) >consensus UUUGCCUACCCCUCGAAAUAAAUCCGUAAAUACCCA____________________UUUUUUCGGACGGAACCCCUU_UUGGCCGGACUUUGUCCGGCGGUCAGAAUGGCA_A ..((((.(((.....................................................((........))......(((((((...))))))))))......)))).. (-14.79 = -15.23 + 0.45)

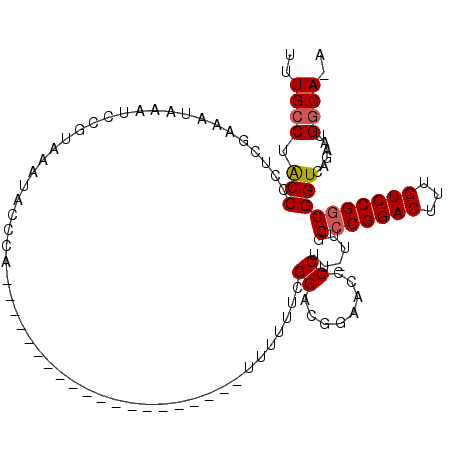
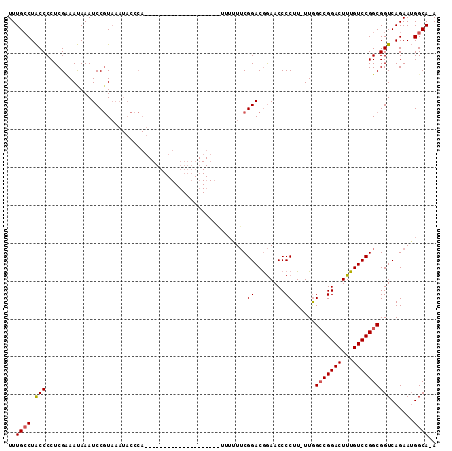
| Location | 1,676,721 – 1,676,838 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -15.61 |
| Consensus MFE | -11.95 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676721 117 - 22407834 AGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGUCCAAAUGGCGAACAACAAAAAUAACAAUUUGCCUACCCCUCGCAAUAAAUCCGUAAAUACCCAA--- .(((((((.......(((.(((((......))))))))............((((.....))))...............)))))))................................--- ( -14.50) >DroSec_CAF1 3180 108 - 1 AGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAAUUUGGCUACCCCUCGUAAUAAAUCCCUU------------ .((............(((.(((((......)))))))).............((((((((....................)))))))).......))............------------ ( -14.15) >DroSim_CAF1 4088 120 - 1 AGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAAUUUGGCUACCCCUCGCAAUAAAUCCGUAAAUACCCACGAA .((............(((.(((((......)))))))).............((((((((....................)))))))).......))........(((........))).. ( -17.15) >DroYak_CAF1 4224 120 - 1 AGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACUGGCCAAAUGGCAAACAACAAAAAUAACAAUUUGCCUGCCCCUCGAAACAAAUCCGUAAAUACCCAUGCA .(((...........(((.(((((......)))))))).............(((.....((((((...............)))))).)))..........................))). ( -16.66) >consensus AGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAAUUUGCCUACCCCUCGCAAUAAAUCCGUAAAUACCCA____ ((((((((.......(((.(((((......))))))))..............(((....)))................)))))).))................................. (-11.95 = -12.20 + 0.25)

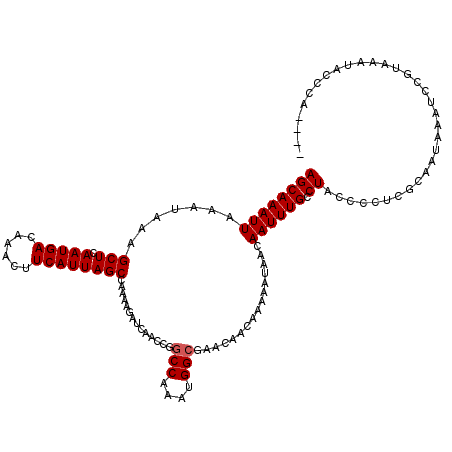
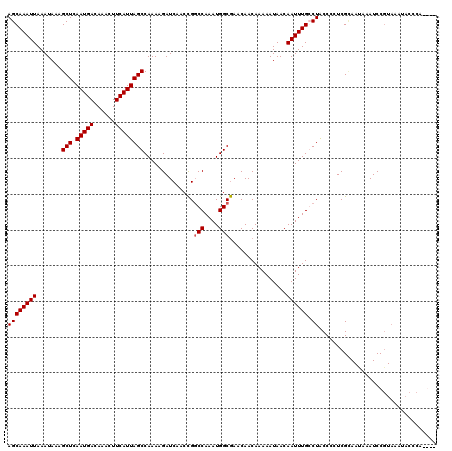
| Location | 1,676,758 – 1,676,872 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -30.46 |
| Energy contribution | -29.90 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676758 114 + 22407834 UUGUUAUUUUUGUUGUUCGCCAUUUGGACGGUUGAUCUUUUGGCUAAUGAAGUUUGUCAUUGAGCUUUAUUUAAUUUGCUGAUGAUGCGAAUGGAAAGAACAACAGAAGGAA------CG ..(((.((((((((((((.(((((((...(((..(....)..)))((((((((((......))))))))))................)))))))...)))))))))))).))------). ( -36.10) >DroSec_CAF1 3208 116 + 1 UUGUUAUUUUUGUUGUUCGCCAUUUGGCCGGUUGAUCUUUUGGCUAAUGAAGUUUGUCAUUGAGCUUUAUUUAAUUUGCUGAUGAUGCGAAU----AGAACAACAGAAGGAAAGGGAUCG ......(((((((((((((((....))).(((..(....)..)))((((((((((......))))))))))..((((((.......))))))----.))))))))))))........... ( -32.30) >DroSim_CAF1 4128 116 + 1 UUGUUAUUUUUGUUGUUCGCCAUUUGGCCGGUUGAUCUUUUGGCUAAUGAAGUUUGUCAUUGAGCUUUAUUUAAUUUGCUGAUGAUGCAAAU----AGAACAACAGAAGGAAAGGGAACG ......(((((((((((((((....))).(((..(....)..)))((((((((((......))))))))))..((((((.......))))))----.))))))))))))....(....). ( -32.80) >DroYak_CAF1 4264 116 + 1 UUGUUAUUUUUGUUGUUUGCCAUUUGGCCAGUUGAUCUUUUGGCUAAUGAAGUUUGUCAUUGAGCUUUAUUUAAUUUGCUGAUGAUGCGAAU----AAAACAACAGAAGGCAAGGGAAUG ((((..((((((((((((.....((((((((........))))))))((((((((......))))))))....((((((.......))))))----.))))))))))))))))....... ( -31.50) >consensus UUGUUAUUUUUGUUGUUCGCCAUUUGGCCGGUUGAUCUUUUGGCUAAUGAAGUUUGUCAUUGAGCUUUAUUUAAUUUGCUGAUGAUGCGAAU____AGAACAACAGAAGGAAAGGGAACG ......((((((((((((((((...(..(....)..)...)))).((((((((((......))))))))))..((((((.......)))))).....))))))))))))........... (-30.46 = -29.90 + -0.56)

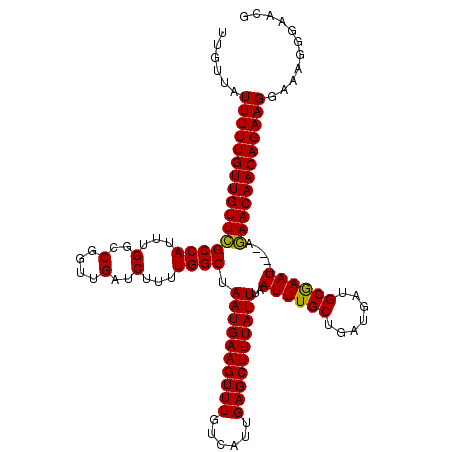
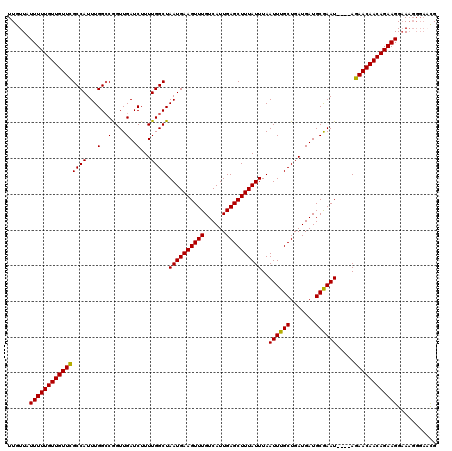
| Location | 1,676,758 – 1,676,872 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.35 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676758 114 - 22407834 CG------UUCCUUCUGUUGUUCUUUCCAUUCGCAUCAUCAGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGUCCAAAUGGCGAACAACAAAAAUAACAA .(------((..((.((((((((...(((((.((.......))............(((.(((((......))))))))..................))))).)))))))).))..))).. ( -21.10) >DroSec_CAF1 3208 116 - 1 CGAUCCCUUUCCUUCUGUUGUUCU----AUUCGCAUCAUCAGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAA ...............((((((((.----..((((.......))............(((.(((((......)))))))).....)).......(((....))))))))))).......... ( -21.00) >DroSim_CAF1 4128 116 - 1 CGUUCCCUUUCCUUCUGUUGUUCU----AUUUGCAUCAUCAGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAA ...............((((((((.----((((((.......))))))........(((.(((((......))))))))..............(((....))))))))))).......... ( -23.40) >DroYak_CAF1 4264 116 - 1 CAUUCCCUUGCCUUCUGUUGUUUU----AUUCGCAUCAUCAGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACUGGCCAAAUGGCAAACAACAAAAAUAACAA ...............((((((((.----..((((.......))............(((.(((((......)))))))).....)).......(((....))))))))))).......... ( -17.90) >consensus CGUUCCCUUUCCUUCUGUUGUUCU____AUUCGCAUCAUCAGCAAAUUAAAUAAAGCUCAAUGACAAACUUCAUUAGCCAAAAGAUCAACCGGCCAAAUGGCGAACAACAAAAAUAACAA ...............((((((((.........((.......))............(((.(((((......))))))))..............(((....))))))))))).......... (-18.48 = -18.35 + -0.13)

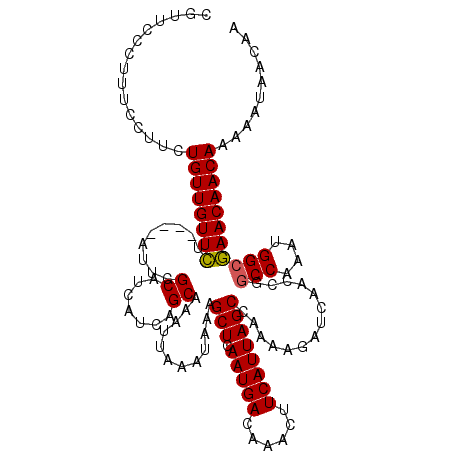
| Location | 1,676,872 – 1,676,992 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -17.61 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.61 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1676872 120 - 22407834 UUUACAGAUUUUACUUUCAAAUUUCAAAAUCGCUGAUACUCUUCUGUAUCAAUAAUUCCUUUUUAAAAUCAUUAACCCAUCUGGAAUCGAACCUUUUUAGGCCUUUGUUUGAUUGCCACC ......(((((((.(((........))).....((((((......))))))............)))))))...........(((((((((((..............)))))))).))).. ( -16.04) >DroSec_CAF1 3324 104 - 1 --------------UUUAAAAUUUG--AAUCGCUGAUACACUUCUGUAUCAUAUAUUCCCUUUUAAAAUCAUUAACCCAUCUGGAAUCGAACCUUUUUAAGCCUUUGUUUGAUUGCCGCC --------------(((((((...(--(((...(((((((....)))))))...))))..)))))))...............((((((((((..............)))))))).))... ( -17.64) >DroSim_CAF1 4244 118 - 1 CUUACAGAUUUUACUUUCAAAUUUG--AAUCGCUGAUACACGUCUGUAUCAUAUGAUCCCUUUUAAAAUCAUUAACCUAUCUGGAAUCGAACCUUUUUAUGGCUUUGUUUGAUUGCCGCC ......(((((((..((((....))--))....(((((((....)))))))............)))))))............((((((((((..............)))))))).))... ( -19.14) >consensus _UUACAGAUUUUACUUUCAAAUUUG__AAUCGCUGAUACACUUCUGUAUCAUAUAUUCCCUUUUAAAAUCAUUAACCCAUCUGGAAUCGAACCUUUUUAAGCCUUUGUUUGAUUGCCGCC .................................((((((......))))))..............................(((((((((((..............)))))))).))).. (-13.83 = -13.61 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:46 2006