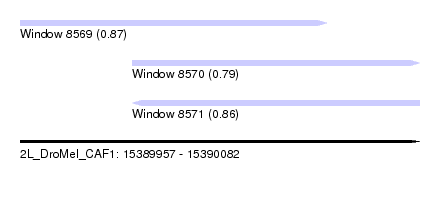
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,389,957 – 15,390,082 |
| Length | 125 |
| Max. P | 0.874522 |

| Location | 15,389,957 – 15,390,053 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -9.22 |
| Energy contribution | -9.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15389957 96 + 22407834 ---UGUCUAAAAUGGCCCAAAAGGCAGCCCCCA-AGUCAAAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUU ---.......(((((((.....((......)).-.(((....)))......((((((((.((((.(((((....)))))))))))))))))..))))))) ( -26.80) >DroVir_CAF1 161036 90 + 1 AACUGUCGAGU-UGGCCCAAAAGGCAGCU-CCA-ACUCAAAUGACACUAACAACAACAAUAAACUACUGUAGACAU-------UUGUUGUUGUUGUUGCU ...((((((((-(((.((....)).....-)))-))))....)))).((((((((((((((((....((....)))-------))))))))))))))).. ( -27.00) >DroWil_CAF1 239542 93 + 1 ---UGUCUAAAAUGGCCCAAAAGGCAGUCGCCACAGUCAAAUGACACUAACAACAACAAUAAACUACUGUUGACAU----UUGUUGCUGUUGUUGCUGGA ---.(((......)))(((...(((....)))...(((....)))..((((((((.(((((((....((....)))----)))))).)))))))).))). ( -22.20) >DroMoj_CAF1 169858 91 + 1 AACUGUCGACUGUGGCCCAAAAGGCAGCU-CCA-AGUCAAAUGACACUAACAACAACAAUAAACUACUGUUGACAU-------UUGUUGUUGCUGUUGUU .......(((((..((((....))..)).-.).-)))).((..(((.(((((((((.....(((....))).....-------))))))))).)))..)) ( -23.20) >DroAna_CAF1 142699 87 + 1 ---UGUCUAAAAUGGCCCAAAAGGCAGCCCCCA-AGUCAAAUGACACUUAAAACAACAAUAAACUACUGUUGACAUACUGUUUUUGUUUU---------U ---.(((......)))...((((((((......-.(((....)))........(((((.........))))).....)))))))).....---------. ( -13.50) >DroPer_CAF1 137430 84 + 1 ---UGUCGAAAAUGGCCCAAAAGGCAGCCCCCG-AGUCAAAUGACACCAGCAGCAACAAUAAACAAAAGGCGACA-------UUUGUUG---GGGCUG-- ---...........(((.....)))((((((..-.(((....)))................((((((.(....).-------)))))))---))))).-- ( -23.30) >consensus ___UGUCGAAAAUGGCCCAAAAGGCAGCCCCCA_AGUCAAAUGACACUAACAACAACAAUAAACUACUGUUGACAU______UUUGUUGUUGUUGCUG_U ...((((.......(((.....)))..........(((....)))..........................))))......................... ( -9.22 = -9.38 + 0.17)

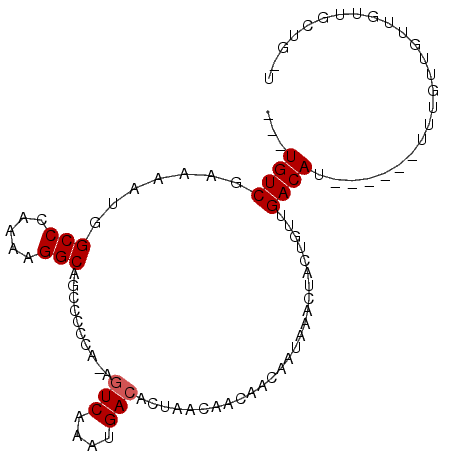

| Location | 15,389,992 – 15,390,082 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -16.31 |
| Energy contribution | -17.37 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15389992 90 + 22407834 AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUUGUUGCCAUUUUGCUCUCUAGUCCAUUCAA ...(((......((((((((.((((.(((((....)))))))))))))))))(((((.......)))))...........)))....... ( -21.60) >DroSec_CAF1 135801 90 + 1 AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUUGUUGCCAUUUUGCUCUCUAGUCCAUUCAA ...(((......((((((((.((((.(((((....)))))))))))))))))(((((.......)))))...........)))....... ( -21.60) >DroSim_CAF1 140723 90 + 1 AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUAGUUGCCAUUUUGCUCUCUAGUCCAUUCAA ...(((......((((((((.((((.(((((....)))))))))))))))))(((((.......)))))...........)))....... ( -21.60) >DroEre_CAF1 142132 90 + 1 AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUUGUUGCCAUUUUGCUCCCCAGUCCAUUCAA ...(((......((((((((.((((.(((((....)))))))))))))))))(((((.......)))))...........)))....... ( -21.60) >DroYak_CAF1 142763 90 + 1 AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUUGUUGCCAUUUGGCUCUCCAGUCCAUUCAA ..(((.......((((((((.((((.(((((....)))))))))))))))))((((.(......(((....)))......).))))))). ( -22.90) >DroAna_CAF1 142734 81 + 1 AAUGACACUUAAAACAACAAUAAACUACUGUUGACAUACUGUUUUUGUUUU---------UGUUGCCAUUUGGCUCUUACGUUCGAAUAC ...(((......(((((.((((((..((.((......)).)).)))))).)---------))))(((....)))......)))....... ( -11.40) >consensus AAUGACACUAACAACAACAAUAAACUACUGUUGACACAGUGUUUUUGUUGUUAUGGCCGUUGUUGCCAUUUUGCUCUCUAGUCCAUUCAA ...(((......((((((((.((((.(((((....)))))))))))))))))(((((.......)))))...........)))....... (-16.31 = -17.37 + 1.06)


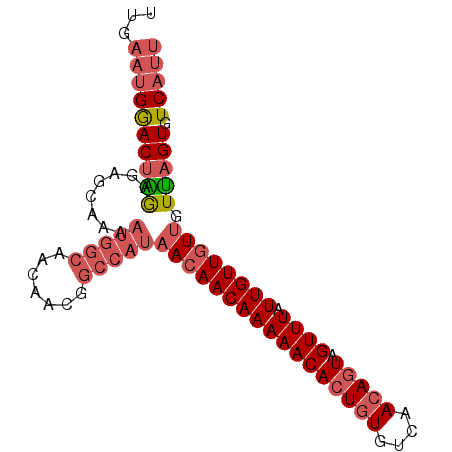
| Location | 15,389,992 – 15,390,082 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.75 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15389992 90 - 22407834 UUGAAUGGACUAGAGAGCAAAAUGGCAACAACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...((((((((((........(((((.......)))))(((((((((((((((((....))))).)))).)))))))).))))).))))) ( -26.50) >DroSec_CAF1 135801 90 - 1 UUGAAUGGACUAGAGAGCAAAAUGGCAACAACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...((((((((((........(((((.......)))))(((((((((((((((((....))))).)))).)))))))).))))).))))) ( -26.50) >DroSim_CAF1 140723 90 - 1 UUGAAUGGACUAGAGAGCAAAAUGGCAACUACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...((((((((((........(((((.......)))))(((((((((((((((((....))))).)))).)))))))).))))).))))) ( -26.50) >DroEre_CAF1 142132 90 - 1 UUGAAUGGACUGGGGAGCAAAAUGGCAACAACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...((((((((((........(((((.......)))))(((((((((((((((((....))))).)))).)))))))).))))).))))) ( -26.10) >DroYak_CAF1 142763 90 - 1 UUGAAUGGACUGGAGAGCCAAAUGGCAACAACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...(((((((((..(.(((...((....))..))))(((((((((((((((((((....))))).)))).)))))))))))))).))))) ( -28.10) >DroAna_CAF1 142734 81 - 1 GUAUUCGAACGUAAGAGCCAAAUGGCAACA---------AAAACAAAAACAGUAUGUCAACAGUAGUUUAUUGUUGUUUUAAGUGUCAUU .........(....).....(((((((...---------((((((...((((((....(((....)))))))))))))))...))))))) ( -12.70) >consensus UUGAAUGGACUAGAGAGCAAAAUGGCAACAACGGCCAUAACAACAAAAACACUGUGUCAACAGUAGUUUAUUGUUGUUGUUAGUGUCAUU ...((((((((((........(((((.......)))))(((((((((((((((((....))))).)))).)))))))).))))).))))) (-19.30 = -20.75 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:12 2006