
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,379,381 – 15,379,483 |
| Length | 102 |
| Max. P | 0.788047 |

| Location | 15,379,381 – 15,379,483 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.29 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -6.07 |
| Energy contribution | -7.29 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15379381 102 - 22407834 GUUUAGUGGAGCACAGCCUCCACGUCCUGCUCCG----CCGACCAACAGCCUCC-GUUC--------UCCAUCCCCUAUCCUCGGCCUCAUCUUCAUCAUCUGA-UGGUCCCAUCC--- .....((((((((..((......))..)))))))----).(((((.(((.....-....--------......((........))...............))).-)))))......--- ( -23.31) >DroPse_CAF1 127643 92 - 1 GUUUAGUGGAGC--------CAGAGCCAGCUCCAAGCCACGAUGAUCAUUCGCA-GUG-----------------CCAGGCACAGCCUCAUCUUCAUCAUUCUC-CGGGCUCAUCCUCG .....(.(((..--------..(((((.((.....))...(((((......((.-(((-----------------(...)))).)).......)))))......-..))))).))).). ( -24.82) >DroSec_CAF1 125230 102 - 1 GUUUAGUGGAGCACUGCCUCCACGUCCUGCUCCA----CCGACCAACAUCCUCC-GUUC--------UCCAUCCCCUAUCCUCGGCCUCAUCUUCAUCAUCUGA-UGGUCCCAUCC--- .....((((((((..((......))..)))))))----).(((((.......((-(...--------...............)))........(((.....)))-)))))......--- ( -21.57) >DroSim_CAF1 129357 102 - 1 GUUUAGUGGAGCACAGCCUCCACGUCCUGCUCCA----CCGACCAACAUCCUCC-GUUC--------UCCAUCCCCUAUCCUCGGCCUCAUCUUCAUCAUCUGA-UGGUCCCAUCC--- .....((((((((..((......))..)))))))----).(((((.......((-(...--------...............)))........(((.....)))-)))))......--- ( -21.77) >DroYak_CAF1 132204 92 - 1 GUUUAGUGGAGCACAGCCUCCACUGCCU---CCA----CUG--------CAUCCGCUCC--------UCCAUCCCCUAUCCUCGGCCUCAUCUUCAUCAUCUGA-UGGUCCCAUCC--- ...((((((((..(((......))).))---)))----)))--------..........--------................((..(((((..........))-)))..))....--- ( -19.40) >DroAna_CAF1 132804 100 - 1 GUUUACUGGAGCACUGACU---------GCUCCU----UCGAGCCACUCCCCUC-GAUCCGAACCGAUCCAUC--CGGUCCUCGGCCUCAUCUUCAUCAUCUGGCCAGUCUCAUCC--- .......((((((.....)---------))))).----(((((........)))-))...((((((.......--))))..))((((...............))))..........--- ( -24.96) >consensus GUUUAGUGGAGCACAGCCUCCACGUCCUGCUCCA____CCGACCAACAUCCUCC_GUUC________UCCAUCCCCUAUCCUCGGCCUCAUCUUCAUCAUCUGA_UGGUCCCAUCC___ .......(((((................)))))..................................................((((...((..........))..))))......... ( -6.07 = -7.29 + 1.22)
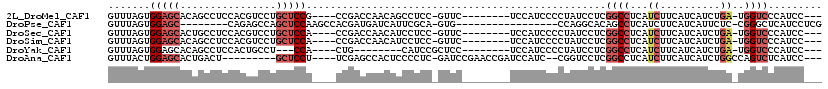

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:02 2006