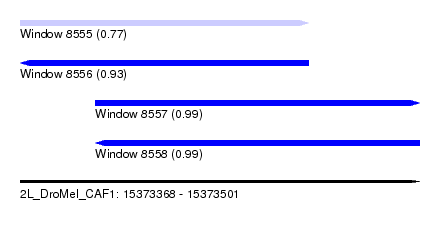
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,373,368 – 15,373,501 |
| Length | 133 |
| Max. P | 0.992240 |

| Location | 15,373,368 – 15,373,464 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15373368 96 + 22407834 UGUAGAUUCUCCCACC-UU------CCAC---AGUUCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA ................-..------.(((---(....))))..........(((.((((((((.((.((((....(((....))).)))).)))))))))).))). ( -28.70) >DroSec_CAF1 119194 96 + 1 UGUUGAUUCUCCCACC-UU------CCAC---AGUUCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA ................-..------.(((---(....))))..........(((.((((((((.((.((((....(((....))).)))).)))))))))).))). ( -28.70) >DroSim_CAF1 123294 96 + 1 UGUAGAUUCUCCCACC-UU------CCAC---AGUUCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA ................-..------.(((---(....))))..........(((.((((((((.((.((((....(((....))).)))).)))))))))).))). ( -28.70) >DroEre_CAF1 125423 96 + 1 UGUAGAUUCUCCCACC-UU------CCAC---AGUUCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGCUGCAUUUGCUGCGUUGCAGCCGCAA ................-..------.(((---(....))))..........(((.((((((((.((.((((....(((....))).)))).)))))))))).))). ( -28.70) >DroYak_CAF1 125416 90 + 1 UCUAGAUUCUCCCACC-UUUCACAUCCAC---AGUUCUGUGACAAUAUUCCUGCCGCUGCAACUGUUGCAACUU------------UUGCUGCGUUGCAGCCGCAA ................-.........(((---(....))))..........(((.((((((((.((.(((....------------.))).)))))))))).))). ( -25.30) >DroAna_CAF1 127139 89 + 1 UGCA-AUUCCACUACCACU------CGACGCGGGUUUUGUGACAAUAUUUCUGCCACUGCAG-UGGUGCAACUUC---------AGUUGCUGCGUUGCAGCCGCAA ....-..............------....((((...(((..((.........(((((....)-))))(((((...---------.)))))...))..))))))).. ( -26.10) >consensus UGUAGAUUCUCCCACC_UU______CCAC___AGUUCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA ...................................................(((.((((((((.((.((((....(((....))).)))).)))))))))).))). (-20.20 = -20.93 + 0.74)

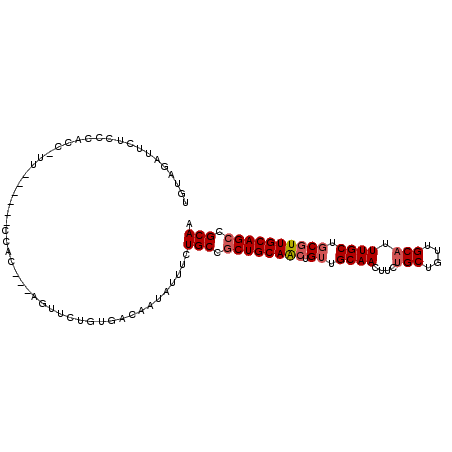

| Location | 15,373,368 – 15,373,464 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.27 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15373368 96 - 22407834 UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGAACU---GUGG------AA-GGUGGGAGAAUCUACA ((((.(((((((((((((...(((....)))...)))))....)))))))).))))........(((((....)---))))------..-.((((......)))). ( -35.60) >DroSec_CAF1 119194 96 - 1 UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGAACU---GUGG------AA-GGUGGGAGAAUCAACA ((((.(((((((((((((...(((....)))...)))))....)))))))).))))........(((((....)---))))------..-(((......))).... ( -33.60) >DroSim_CAF1 123294 96 - 1 UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGAACU---GUGG------AA-GGUGGGAGAAUCUACA ((((.(((((((((((((...(((....)))...)))))....)))))))).))))........(((((....)---))))------..-.((((......)))). ( -35.60) >DroEre_CAF1 125423 96 - 1 UUGCGGCUGCAACGCAGCAAAUGCAGCAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGAACU---GUGG------AA-GGUGGGAGAAUCUACA ((((.(((((((((((.....))).(((((....)))))....)))))))).))))........(((((....)---))))------..-.((((......)))). ( -37.50) >DroYak_CAF1 125416 90 - 1 UUGCGGCUGCAACGCAGCAA------------AAGUUGCAACAGUUGCAGCGGCAGGAAUAUUGUCACAGAACU---GUGGAUGUGAAA-GGUGGGAGAAUCUAGA ((((.(((((((((((((..------------..)))))....)))))))).))))........(((((....)---)))).......(-(((......))))... ( -30.40) >DroAna_CAF1 127139 89 - 1 UUGCGGCUGCAACGCAGCAACU---------GAAGUUGCACCA-CUGCAGUGGCAGAAAUAUUGUCACAAAACCCGCGUCG------AGUGGUAGUGGAAU-UGCA ((((....)))).((((((((.---------...))))).(((-((((.(((((((.....)))))))......(((....------.))))))))))...-))). ( -33.00) >consensus UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGAACU___GUGG______AA_GGUGGGAGAAUCUACA ((((.(((((((((((((...(((....)))...)))))....)))))))).))))...................................((((......)))). (-25.44 = -26.27 + 0.82)

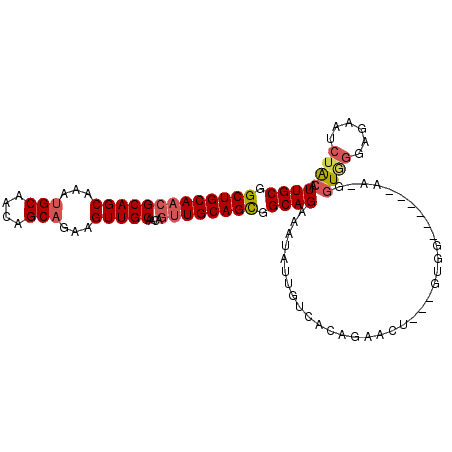
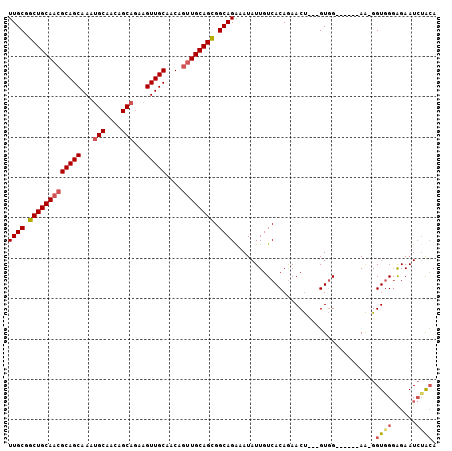
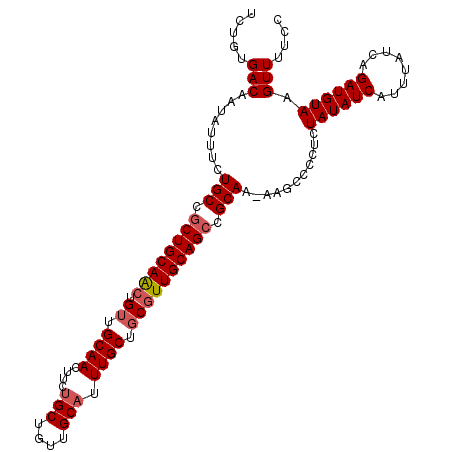
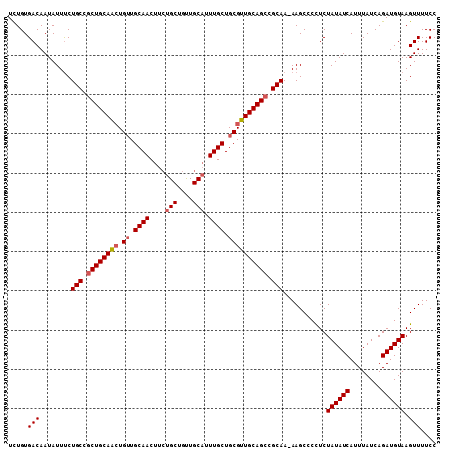
| Location | 15,373,393 – 15,373,501 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -24.30 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15373393 108 + 22407834 UCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA-AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.((((....(((....))).)))).)))))))))).))).-.........((((((........)))))).))).... ( -29.40) >DroSec_CAF1 119219 108 + 1 UCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA-AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.((((....(((....))).)))).)))))))))).))).-.........((((((........)))))).))).... ( -29.40) >DroSim_CAF1 123319 108 + 1 UCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA-AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.((((....(((....))).)))).)))))))))).))).-.........((((((........)))))).))).... ( -29.40) >DroEre_CAF1 125448 108 + 1 UCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGCUGCAUUUGCUGCGUUGCAGCCGCAA-AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.((((....(((....))).)))).)))))))))).))).-.........((((((........)))))).))).... ( -29.40) >DroYak_CAF1 125447 96 + 1 UCUGUGACAAUAUUCCUGCCGCUGCAACUGUUGCAACUU------------UUGCUGCGUUGCAGCCGCAA-AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.(((....------------.))).)))))))))).))).-.........((((((........)))))).))).... ( -26.00) >DroAna_CAF1 127167 99 + 1 UUUGUGACAAUAUUUCUGCCACUGCAG-UGGUGCAACUUC---------AGUUGCUGCGUUGCAGCCGCAAAAAGCUCUUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .(((..((.........(((((....)-))))(((((...---------.)))))...))..)))......((((((....((((((........)))))))))))).. ( -24.30) >consensus UCUGUGACAAUAUUUCUGCCGCUGCAACUGUUGCAACUUCUGCUGUUGCAUUUGCUGCGUUGCAGCCGCAA_AAGCCCCUCUAUAUCAUUUAUCAGAUGUAAGUUUUCC .....(((........(((.((((((((.((.((((....(((....))).)))).)))))))))).)))...........((((((........)))))).))).... (-24.30 = -25.03 + 0.74)

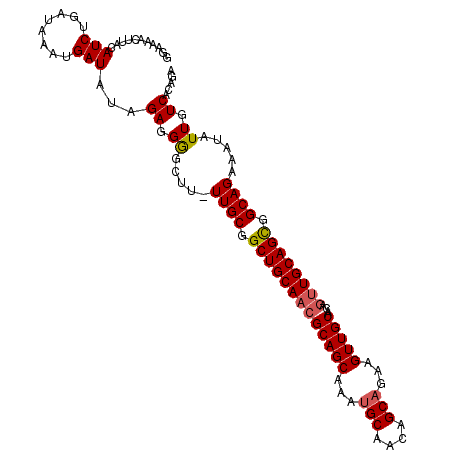
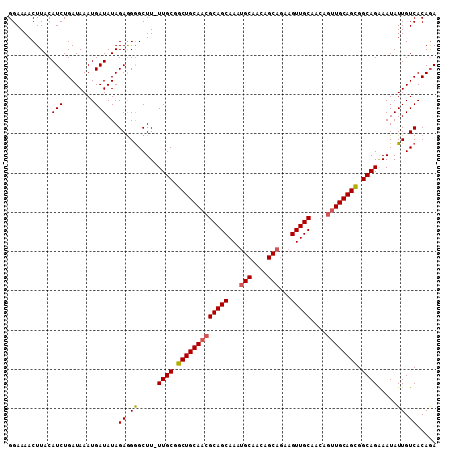
| Location | 15,373,393 – 15,373,501 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -27.65 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15373393 108 - 22407834 GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU-UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGA ............(((((((.....))).((.((..((-((((.(((((((((((((...(((....)))...)))))....)))))))).))))))...)).)).)))) ( -34.40) >DroSec_CAF1 119219 108 - 1 GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU-UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGA ............(((((((.....))).((.((..((-((((.(((((((((((((...(((....)))...)))))....)))))))).))))))...)).)).)))) ( -34.40) >DroSim_CAF1 123319 108 - 1 GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU-UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGA ............(((((((.....))).((.((..((-((((.(((((((((((((...(((....)))...)))))....)))))))).))))))...)).)).)))) ( -34.40) >DroEre_CAF1 125448 108 - 1 GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU-UUGCGGCUGCAACGCAGCAAAUGCAGCAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGA ............(((((((.....))).((.((..((-((((.(((((((((((.....))).(((((....)))))....)))))))).))))))...)).)).)))) ( -36.30) >DroYak_CAF1 125447 96 - 1 GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU-UUGCGGCUGCAACGCAGCAA------------AAGUUGCAACAGUUGCAGCGGCAGGAAUAUUGUCACAGA ............(((((((.....))).((.((..((-((((.(((((((((((((..------------..)))))....)))))))).))))))...)).)).)))) ( -30.10) >DroAna_CAF1 127167 99 - 1 GGAAAACUUACAUCUGAUAAAUGAUAUAGAAGAGCUUUUUGCGGCUGCAACGCAGCAACU---------GAAGUUGCACCA-CUGCAGUGGCAGAAAUAUUGUCACAAA ...........(((........)))...((.((..(((((((.((((((.....(((((.---------...)))))....-.)))))).)))))))..)).))..... ( -26.00) >consensus GGAAAACUUACAUCUGAUAAAUGAUAUAGAGGGGCUU_UUGCGGCUGCAACGCAGCAAAUGCAACAGCAGAAGUUGCAACAGUUGCAGCGGCAGAAAUAUUGUCACAGA ...........(((........)))...((.((.....((((.(((((((((((((...(((....)))...)))))....)))))))).)))).....)).))..... (-27.65 = -28.08 + 0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:00 2006