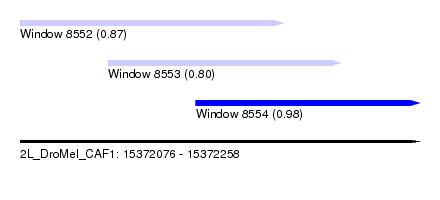
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,372,076 – 15,372,258 |
| Length | 182 |
| Max. P | 0.979334 |

| Location | 15,372,076 – 15,372,196 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.59 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
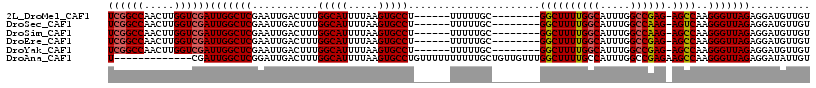
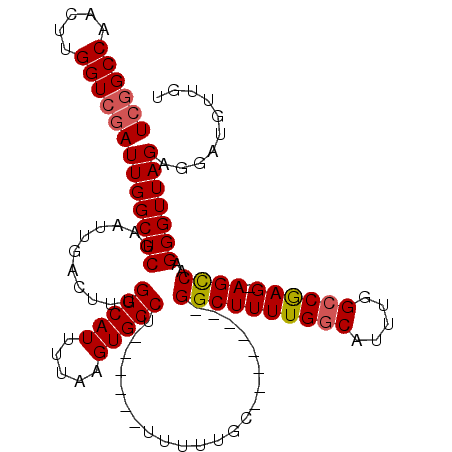
>2L_DroMel_CAF1 15372076 120 + 22407834 AAAUGUGUGUCUAUAUGGCCUGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. ( -39.24) >DroSec_CAF1 117899 120 + 1 AAAUGUGUGUCUAUAUGGCCAGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. ( -39.74) >DroSim_CAF1 122003 120 + 1 AAAUGUGUGUCUAUAUGGCCAGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. ( -39.74) >DroEre_CAF1 124088 120 + 1 AAAUGUGUGUCUAUAUGGCUUGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. ( -37.24) >DroYak_CAF1 124049 120 + 1 AAAUGUGUGUCUAUAUGGCUUGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. ( -37.24) >DroAna_CAF1 125772 107 + 1 AAAUGUGUUUCUAUAUGGCCUGCUUAUUACCUAAUUUCAUUCGGCCCGUUGCAUUGACAGGCGUUUGCUCAUUCGCCCCGU-------------CGAUUGGCUCGGAUUGACUUUGGCAU ..............................((((..((((((((((......((((((.((((..(....)..))))..))-------------)))).))).)))).)))..))))... ( -22.50) >consensus AAAUGUGUGUCUAUAUGGCCUGCUUAUUACCUAAUUUCAUUGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAU ((((((.((((...((((((.(((((..............)))))..))).))).)))).))))))((((((((((((.(((((((.....))))))).))).)))).))).....)).. (-32.96 = -33.59 + 0.63)

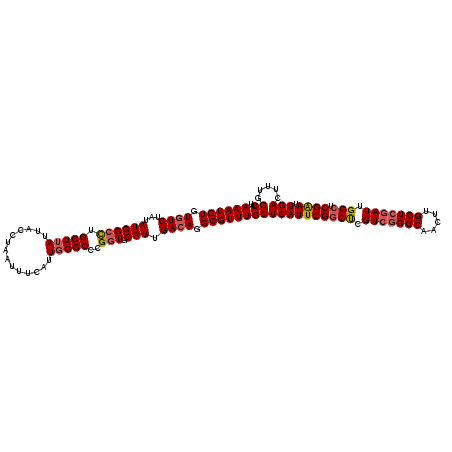
| Location | 15,372,116 – 15,372,222 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15372116 106 + 22407834 UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... ( -35.20) >DroSec_CAF1 117939 106 + 1 UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... ( -35.20) >DroSim_CAF1 122043 106 + 1 UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... ( -35.20) >DroEre_CAF1 124128 106 + 1 UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... ( -35.20) >DroYak_CAF1 124089 106 + 1 UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... ( -35.20) >consensus UGGGCCCGGUGCAUUGACAGGCGUUUGCUCAUUCGGCUCGUCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCUUUUUUGCGGCUUUUG .(((((....((........))....((((((((((((.(((((((.....))))))).))).)))).)))....(((((.....)))))......)))))))... (-35.20 = -35.20 + -0.00)

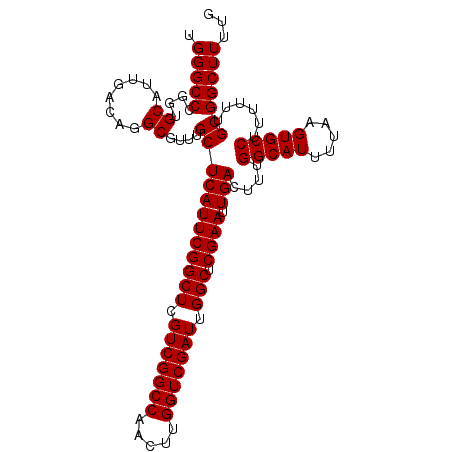

| Location | 15,372,156 – 15,372,258 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -30.65 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15372156 102 + 22407834 UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU------UUUUUGC--------GGCUUUUGGCAUUUGGCCGAG-AGCCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....))))).------.......--------((((((((((.....))))))-))))..))))))).......... ( -38.80) >DroSec_CAF1 117979 102 + 1 UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU------UUUUUGC--------GGCUUUUGGCAUUUGGCCAAG-AGUCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....))))).------.......--------((((((((((.....))))))-))))..))))))).......... ( -36.40) >DroSim_CAF1 122083 102 + 1 UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU------UUUUUGC--------GGCUUUUGGCAUUUGGCCAAG-AGCCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....))))).------.......--------((((((((((.....))))))-))))..))))))).......... ( -39.10) >DroEre_CAF1 124168 102 + 1 UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU------UUUUUGC--------GGCUUUUGGCAUUUGGCCGAG-AGCCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....))))).------.......--------((((((((((.....))))))-))))..))))))).......... ( -38.80) >DroYak_CAF1 124129 102 + 1 UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU------UUUUUGC--------GGCUUUUGGCAUUUGGCCGAG-AGCCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....))))).------.......--------((((((((((.....))))))-))))..))))))).......... ( -38.80) >DroAna_CAF1 125852 104 + 1 U-------------CGAUUGGCUCGGAUUGACUUUGGCAUUUUAAGUGCCUGUUUUUUUUUUGCUGUUGUUUGGCUUUUGCCAUUUGGCCGAGAAGCCAAGGGUUAGAGGAUAUUGU (-------------(..((((((((((..(((...(((((.....))))).)))..)))...........((((((((((((....)))..))))))))))))))))..))...... ( -29.20) >consensus UCGGCCAACUUGGUCGAUUGGCUCGAAUUGACUUUGGCAUUUUAAGUGCCU______UUUUUGC________GGCUUUUGGCAUUUGGCCGAG_AGCCAAGGGUUAGAGGAUGUUGU ((((((.....))))))(((((((...........(((((.....)))))......................((((((((((.....)))))).))))..))))))).......... (-30.65 = -30.92 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:56 2006