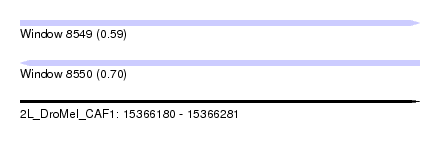
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,366,180 – 15,366,281 |
| Length | 101 |
| Max. P | 0.701022 |

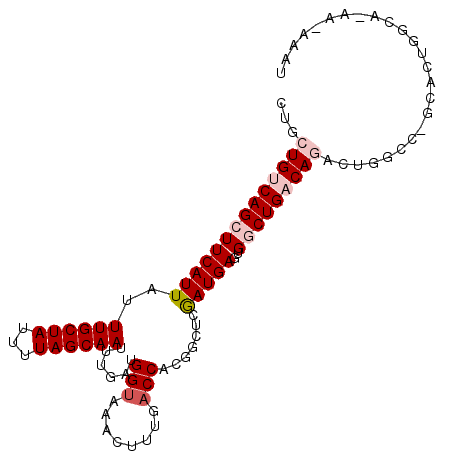
| Location | 15,366,180 – 15,366,281 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.78 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15366180 101 + 22407834 AUUUUUUAUGCCAGUGC-AGACAGUCUGUCAGCCCCUCAUCGAGCCGUGGUCAAAGUUUACCAUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCAG ........(((....))-)....(.((((((((...((((......(((((........)))))....((((((...))))))...)))).))))))))).. ( -24.20) >DroPse_CAF1 114727 82 + 1 ----------------CGGGC---CAUGACAGGCACUCAUCGAGCCGUGGUCAAAGUUUACCUUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCA- ----------------..(((---(((....(((.(.....).)))))))))...(((...(((((..((((((...))))))....)))))..)))....- ( -20.30) >DroSim_CAF1 115843 100 + 1 AUUU-UUCUGCCAGUGC-ACACAGUCUGUCAGCCACUCAUCGAGCCGUGGUCAAAGUUUACCAUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCAG ....-...(((....))-)....(.((((((((...((((......(((((........)))))....((((((...))))))...)))).))))))))).. ( -23.70) >DroYak_CAF1 118270 100 + 1 AUUU-UCCUGCCAGUGC-GGACAUUCUGUCAGCCACUCAUCGAGACGUGGUCAAAGUUUACCAUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCAG ....-..((((....))-)).....((((((((...((((......(((((........)))))....((((((...))))))...)))).))))))))... ( -25.40) >DroAna_CAF1 119975 100 + 1 AUUU-UUUUCUCAGUGC-AGCCCGUCUGUCAGCCACUCAUUGAGCCGUGGUCAAAGUUUGCCAUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCAG ....-............-.....(.((((((((...((((((....(((((........)))))....((((((...)))))).)))))).))))))))).. ( -25.90) >DroPer_CAF1 114712 82 + 1 ----------------CGGGC---CAUGACAGGCACUCAUCGAGCCGUGGUCAAAGUUUACCUUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCG- ----------------..(((---(((....(((.(.....).)))))))))...(((...(((((..((((((...))))))....)))))..)))....- ( -20.30) >consensus AUUU_UU_UGCCAGUGC_AGACAGUCUGUCAGCCACUCAUCGAGCCGUGGUCAAAGUUUACCAUCAAAUUGCUAAAAUAGCAAAUAAUGAAGCUGACAGCAG .........................((((((((...((((......(((((........)))))....((((((...))))))...)))).))))))))... (-15.59 = -16.78 + 1.20)

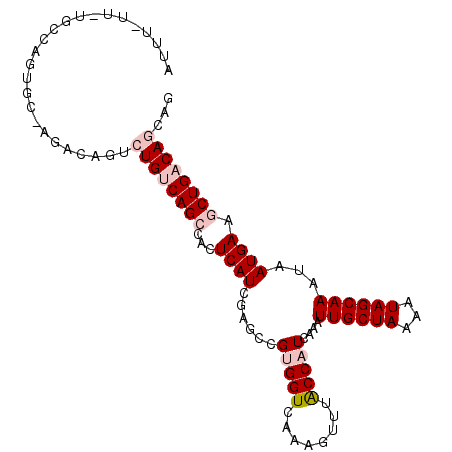
| Location | 15,366,180 – 15,366,281 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15366180 101 - 22407834 CUGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAUGGUAAACUUUGACCACGGCUCGAUGAGGGGCUGACAGACUGUCU-GCACUGGCAUAAAAAAU ...(((((((((((....(((((((((..(....)..)))))))))(.(((((....).)))).).)))))))))))..((((.-.....))))........ ( -30.30) >DroPse_CAF1 114727 82 - 1 -UGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAAGGUAAACUUUGACCACGGCUCGAUGAGUGCCUGUCAUG---GCCCG---------------- -....((((.(((((....((((((...))))))..)))))........((((...(((.(.....).))).))))))---))...---------------- ( -20.40) >DroSim_CAF1 115843 100 - 1 CUGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAUGGUAAACUUUGACCACGGCUCGAUGAGUGGCUGACAGACUGUGU-GCACUGGCAGAA-AAAU ...((((((((((((((..((((((...)))))).(((.((((........)))))))...)))))..))))))))).((((..-......))))..-.... ( -28.80) >DroYak_CAF1 118270 100 - 1 CUGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAUGGUAAACUUUGACCACGUCUCGAUGAGUGGCUGACAGAAUGUCC-GCACUGGCAGGA-AAAU ...((((((((.(((...(((((((((..(....)..)))))))))...))).(((.((.....)))))))))))))....(((-((....)).)))-.... ( -30.00) >DroAna_CAF1 119975 100 - 1 CUGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAUGGCAAACUUUGACCACGGCUCAAUGAGUGGCUGACAGACGGGCU-GCACUGAGAAAA-AAAU (((((((((((((((...(((((((((..(....)..)))))))))...))).....((((...))))))))))))).)))...-............-.... ( -30.70) >DroPer_CAF1 114712 82 - 1 -CGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAAGGUAAACUUUGACCACGGCUCGAUGAGUGCCUGUCAUG---GCCCG---------------- -....((((.(((((....((((((...))))))..)))))........((((...(((.(.....).))).))))))---))...---------------- ( -20.40) >consensus CUGCUGUCAGCUUCAUUAUUUGCUAUUUUAGCAAUUUGAUGGUAAACUUUGACCACGGCUCGAUGAGUGGCUGACAGACUGGCC_GCACUGGCA_AA_AAAU ...((((((((((((((..((((((...))))))......(((........))).......)))))..)))))))))......................... (-18.74 = -19.77 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:52 2006