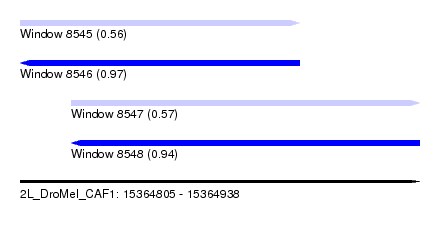
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,364,805 – 15,364,938 |
| Length | 133 |
| Max. P | 0.973969 |

| Location | 15,364,805 – 15,364,898 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15364805 93 + 22407834 -----------------CUCGAGACUUGAGAGGUAGGUUCACCUGUUUAUGCCAGAUUACUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGA---- -----------------.....((((((..(((((((....)))...((((((((....))))))))...........))))((((((....))))))...)).))))..---- ( -28.70) >DroEre_CAF1 117062 100 + 1 -----CUGGA-----GACUUGAGACUUGAGAGGUAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCAUCUGCGCAAGCUUUUGCGCCCACGCAGUGGA---- -----..(..-----..).(((...((((..(.((((....))))..((((((((....)))))))))..))))..)))...((((((....))))))((((...)))).---- ( -28.20) >DroWil_CAF1 184646 84 + 1 ------------------------------GGCGAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUUGGCCAU ------------------------------(((.(((....)))...((((((((....))))))))..........((.(((((...((....)).....))))).))))).. ( -28.60) >DroYak_CAF1 116796 105 + 1 -----UCGGAUUCGGAGCUUGAGACUUGAGAGGUAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUAGA---- -----........((.((..(((.((((....((((((.........((((((((....)))))))).(((....))))))))).)))))))..))..))..........---- ( -31.50) >DroAna_CAF1 118685 95 + 1 --------GAGUCG---GUGGAGUCUUCGGU----GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGC---- --------((.(.(---((((.......(((----((((.....(((((((((((....)))))))).)))...))))))).((((((....)))))))))).)).))..---- ( -33.20) >DroPer_CAF1 113495 106 + 1 CGCAGUCGCAGUCGGAGUCGGAGUCUUCGGA----GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGA---- ...((((((((((((((((((..((....))----((....))........)).)))))).))).)))))))........(((((..(((......)))..)))))....---- ( -37.90) >consensus ________GA_______CUGGAGACUUGAGAGGUAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGA____ ..........................................((((.((((((((....))))))))...............((((((....))))))....))))........ (-21.32 = -21.18 + -0.14)


| Location | 15,364,805 – 15,364,898 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -33.67 |
| Energy contribution | -34.03 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15364805 93 - 22407834 ----UCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGUAAUCUGGCAUAAACAGGUGAACCUACCUCUCAAGUCUCGAG----------------- ----(((((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....)))............)))).----------------- ( -38.20) >DroEre_CAF1 117062 100 - 1 ----UCCACUGCGUGGGCGCAAAAGCUUGCGCAGAUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUACCUCUCAAGUCUCAAGUC-----UCCAG----- ----....(((((..(((......)))..)))))..(((((.......(((((((....)))))))....(((....))).......))))).......-----.....----- ( -33.50) >DroWil_CAF1 184646 84 - 1 AUGGCCAACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUCGCC------------------------------ ..(((...(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))).)))------------------------------ ( -37.50) >DroYak_CAF1 116796 105 - 1 ----UCUACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUACCUCUCAAGUCUCAAGCUCCGAAUCCGA----- ----....(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))).............................----- ( -35.70) >DroAna_CAF1 118685 95 - 1 ----GCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC----ACCGAAGACUCCAC---CGACUC-------- ----....(((((..(((......)))..)))))(((((....)))))(((((((....))))))).....((((....----...........)))---).....-------- ( -36.06) >DroPer_CAF1 113495 106 - 1 ----UCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC----UCCGAAGACUCCGACUCCGACUGCGACUGCG ----(((.(((((..(((......)))..))))).)))......(((((((((((....)))))))....(((....))----).....................))))..... ( -41.70) >consensus ____UCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUACCUCUCAAGUCUCAAG_______UC________ ........(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))).................................. (-33.67 = -34.03 + 0.36)

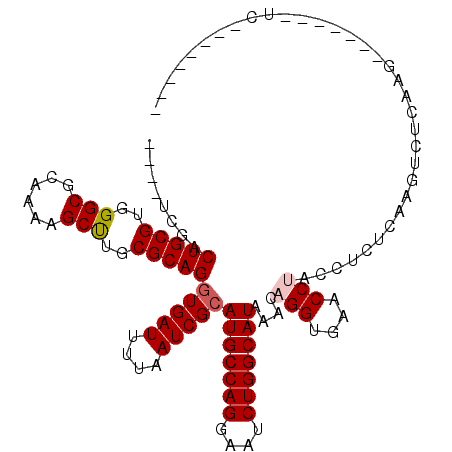
| Location | 15,364,822 – 15,364,938 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -24.75 |
| Energy contribution | -24.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
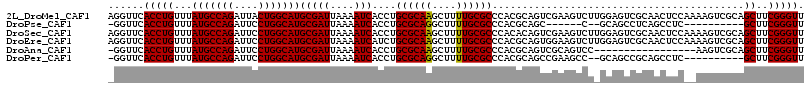
>2L_DroMel_CAF1 15364822 116 + 22407834 AGGUUCACCUGUUUAUGCCAGAUUACUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUCCAAAAGUCGCAGCUUCGGGUU ......(((((...(((((((....)))))))((((((....((..(((((...((....)).....)))))..))....(((((((....))))))).)))))).....))))). ( -40.90) >DroPse_CAF1 113569 97 + 1 -GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGC------C--GCAGCCUCAGCCUC----------GCUUCGGGUU -.....(((((...(((((((....)))))))((((..........(((((..(((......)))..))))).------.--((.......)).))----------))..))))). ( -32.90) >DroSec_CAF1 110819 116 + 1 AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACACAGUCGAAGUCUUGGAGUCGCAACUCCAAAAGUCGCAGCUUCGGGUU ......(((((...(((((((....)))))))((((((..........((((((....))))))................(((((((....))))))).)))))).....))))). ( -38.40) >DroEre_CAF1 117086 116 + 1 AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCAUCUGCGCAAGCUUUUGCGCCCACGCAGUGGAAGUCUUGGAGUCGCAACUCCAAAAGUCGCAGCUUCGGGUU ......(((((...(((((((....)))))))((((((....(((.(((((...((....)).....)))))))).....(((((((....))))))).)))))).....))))). ( -41.40) >DroAna_CAF1 118705 98 + 1 -GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGCAGUCC-----------------AAGUCGCAGCUUCGGGUU -.....(((((...(((((((....)))))))((((((..........((....))..(((((....))))).........-----------------.)))))).....))))). ( -31.70) >DroPer_CAF1 113526 103 + 1 -GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGAAGCC--GCAGCCGCAGCCUC----------GCUUCGGGUU -.....(((((...(((((((....)))))))((((..........((((((.((((((.((((....)).)).))))))--...).)))))..))----------))..))))). ( -37.00) >consensus _GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCU_GGAGUCGCAACUCC__AAGUCGCAGCUUCGGGUU ......(((((...(((((((....)))))))(((((....)))....((((((....))))))..........................................))..))))). (-24.75 = -24.53 + -0.22)
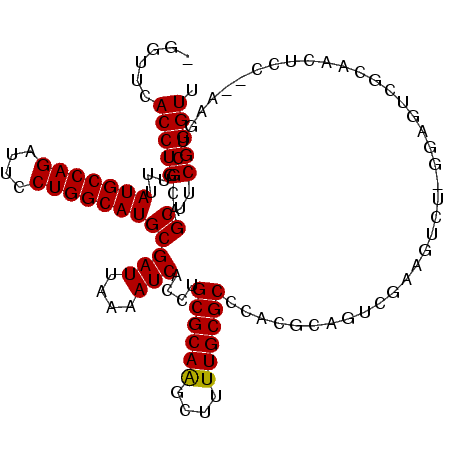

| Location | 15,364,822 – 15,364,938 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
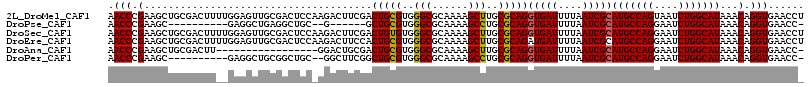
>2L_DroMel_CAF1 15364822 116 - 22407834 AACCCGAAGCUGCGACUUUUGGAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGUAAUCUGGCAUAAACAGGUGAACCU ....(((((.......(((((((((....)))))))))))))).(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))) ( -49.21) >DroPse_CAF1 113569 97 - 1 AACCCGAAGC----------GAGGCUGAGGCUGC--G------GCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC- .(((.(..((----------(((((....))).(--(------.(((((..(((......)))..))))).)).......))))(((((((....)))))))...).))).....- ( -42.70) >DroSec_CAF1 110819 116 - 1 AACCCGAAGCUGCGACUUUUGGAGUUGCGACUCCAAGACUUCGACUGUGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU ....(((((.......(((((((((....)))))))))))))).(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))) ( -47.01) >DroEre_CAF1 117086 116 - 1 AACCCGAAGCUGCGACUUUUGGAGUUGCGACUCCAAGACUUCCACUGCGUGGGCGCAAAAGCUUGCGCAGAUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU ...........((((.(((((((((....)))))))))....(((((((..(((......)))..))))).)).......))))(((((((....)))))))....(((....))) ( -46.80) >DroAna_CAF1 118705 98 - 1 AACCCGAAGCUGCGACUU-----------------GGACUGCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC- .(((.(.....((((.((-----------------(((...((.(((((..(((......)))..))))).))..)))))))))(((((((....)))))))...).))).....- ( -38.30) >DroPer_CAF1 113526 103 - 1 AACCCGAAGC----------GAGGCUGCGGCUGC--GGCUUCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC- .(((.(...(----------((((((((....))--))))))).(((((..(((......)))..)))))(((((....)))))(((((((....)))))))...).))).....- ( -50.00) >consensus AACCCGAAGCUGCGACUU__GGAGUUGCGACUCC_AGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC_ .(((.(......................................(((((..(((......)))..)))))(((((....)))))(((((((....)))))))...).)))...... (-33.78 = -33.58 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:50 2006