
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,307,853 – 15,307,951 |
| Length | 98 |
| Max. P | 0.710782 |

| Location | 15,307,853 – 15,307,951 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.57 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -6.36 |
| Energy contribution | -6.94 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15307853 98 + 22407834 AAACCAGCCGAAAGAUG-------CACAAAAGGCAUUUAGUGGCACUGUGGAAAA----AAUUCUAGAAAUAUAGAAAAUCUUAUGCC---AAUAA-------UAGAAUGUAG-AUAAAU ...(((((((..(((((-------(.......))))))..))))....)))....----..(((((......))))).(((((((...---.....-------....))).))-)).... ( -16.90) >DroSec_CAF1 54777 105 + 1 AAACCAGCCGAAAGAUGGAAGAUGCAUAAAAGGCAUUUAGGGGCACUGUGGAAAA----AAUUCUAGAAUUAUAGAAACUCUUAAGAC---GAUAA-------AAGAAUGUAG-AUAAAU ...(((..(....).)))..(((((.......)))))((((((..(....)....----..(((((......))))).))))))....---.....-------..........-...... ( -13.70) >DroSim_CAF1 55509 102 + 1 AAACCAGCCGAAAGAUGGAAGAUGCA---AAGGCAUUUAGGGGCACUGUGGAAAA----AAUUCUAGAAUUAUAGAAACCCUUAAGAC---GAUAA-------AAGAAUGUAG-AUAAAU ...(((..(....).)))..(((((.---...)))))((((((..(....)....----..(((((......))))).))))))....---.....-------..........-...... ( -17.00) >DroEre_CAF1 58753 97 + 1 AAACCAGCCGAAAGAUG-------CAUAAAAGGCAUUUAGUGGCACUGUGGGAAA----AGUUCUACAAUUAUAAAAACUACCAAGCU---GAUAC-------AAGGAUUUAU-UUAAA- ...((.((((..(((((-------(.......))))))..))))..(((((((..----..)))))))....................---.....-------..))......-.....- ( -20.00) >DroYak_CAF1 57314 91 + 1 AAACCAGCCAAAAGAUG-------CAUAAAAGGCAUUUUUUGGCACUGUGGAAAA----AAUUAUGAAAUUAUAGAAGCUCCCAG----------A-------AGGGAUUUAGCUUAAA- ...((((((((((((((-------(.......))))))))))))....)))....----................((((((((..----------.-------.))))....))))...- ( -26.20) >DroAna_CAF1 60324 111 + 1 AAACCAGCCGAAAGAGG-------GCCAAAAUGCAUUUAGCUGCACAGUGGGAAAUUAAUAUUCCAGGUUCCUGGA-UUUCUUAUAUUUUGAAUAAGGAAGUCAAGAAUAAAG-UUUAAA ...((((.(....).((-------(((....((((......))))...(((((........)))))))))))))).-((((((((.......)))))))).............-...... ( -22.80) >consensus AAACCAGCCGAAAGAUG_______CAUAAAAGGCAUUUAGUGGCACUGUGGAAAA____AAUUCUAGAAUUAUAGAAACUCUUAAGAC___GAUAA_______AAGAAUGUAG_AUAAAU ...(((((((..(((((................)))))..))))....)))..........(((((......)))))........................................... ( -6.36 = -6.94 + 0.58)

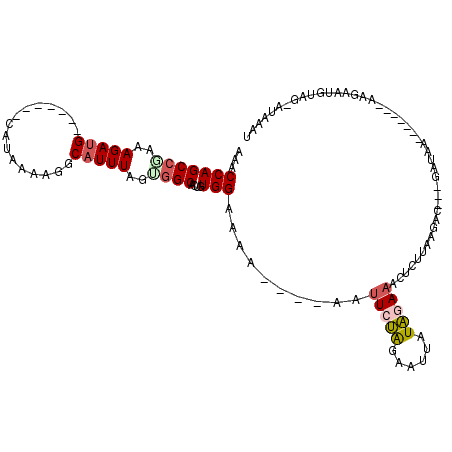
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:41 2006