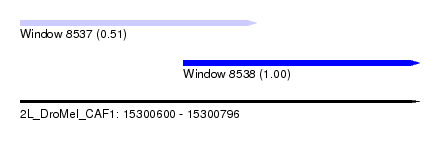
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,300,600 – 15,300,796 |
| Length | 196 |
| Max. P | 0.999468 |

| Location | 15,300,600 – 15,300,716 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15300600 116 + 22407834 AAGGCAAAGGAGCUGGAGUUUGGGCUCGGAGUAGGAGUUGAAGCUGAGCUGAGCAGAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC---- ..(((......(((..(((((..((((((..(((.........)))..)))))).)))))..))).....)))(.((.(((...(((((......)))))..))).)).)......---- ( -41.50) >DroSec_CAF1 47552 112 + 1 AAGGCAAAGGAGCUGGAGUUUGGGCUCGGAGUAGGAGUAGGAGUUG----GAGCUGAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC---- ..(((.......(((..((((.(((((((..(..(........)..----)..))))))).))))..)))..((.((.(((...(((((......)))))..))).)).))..)))---- ( -37.60) >DroSim_CAF1 48350 112 + 1 AAGGCAAAGGAGCUGGAGUUUGGGCCCGGAGUAGGAGUAGGAGUUG----GAGCUGAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGCAGCGAAGUC---- ...((......((((((((((.(((((............)).))).----)))))..((((...))))...)))))(.(((...(((((......)))))..))).).))......---- ( -36.50) >DroEre_CAF1 51661 105 + 1 AAGGCAA-GGAGUUGGAGC-UGGGCUCGGAGUAGGAGUUGGAGCUG---------GAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGUAGUGAAGUU---- ..(((..-(..(((..(((-(.(((((.((.......)).))))).---------).)))..)))..)..)))(.((.(((...(((((......)))))..))).)).)......---- ( -33.50) >DroYak_CAF1 50074 106 + 1 AAGGCAAUGGAGUUGGAGC-UGGGUUCGAGGUAGGAGUUGGUGCUG---------GAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGUAGUGAAGUC---- ..(((.(((........((-..(.(((......))).)..))((((---------(.((((...)))).((((((........))))))....)))))))).)))...........---- ( -32.30) >DroAna_CAF1 52972 96 + 1 AAGGUAAAGGCACUGGAGUU-GGCUUU----------UUGAUGUAG---------GAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCCUGGCC----CGAAGCCUAAC .((((...(((.((((..(.-.(((((----------(......))---------))))..)..)..))))))..((((.(((..((((......))))))).))----))..))))... ( -36.70) >consensus AAGGCAAAGGAGCUGGAGUUUGGGCUCGGAGUAGGAGUUGGAGCUG_________GAGCUGGAGCAGCAGGCCAGUGGGACAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC____ ..(((...((..((((.((((.(((((............................))))).))))....((((((........))))))....))))))...)))............... (-25.42 = -25.17 + -0.25)
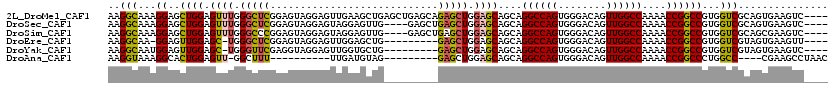
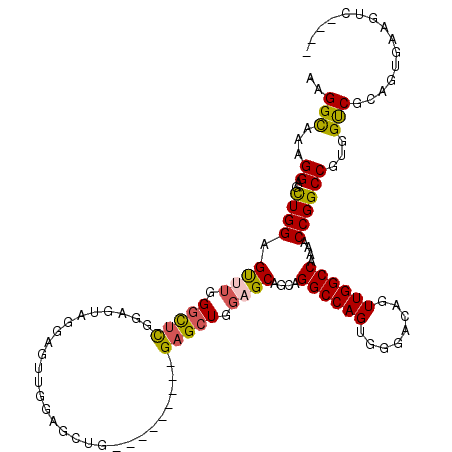
| Location | 15,300,680 – 15,300,796 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -33.85 |
| Energy contribution | -33.47 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15300680 116 + 22407834 CAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC----GGGGCUCUCGCACAGGCCAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((.....((((....))))..(((((((.----...))).))))...)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... ( -34.20) >DroSec_CAF1 47628 116 + 1 CAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC----GAGGCUCUCGCACAGGCCAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((.....((((....))))..(((((((.----...))).))))...)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... ( -34.20) >DroSim_CAF1 48426 116 + 1 CAGUUGGCCAAAACCGGCCGUGGUCGCAGCGAAGUC----GAGGCUCUCGCACAGGCCAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((.....((((....))))..(((((((.----...))).))))...)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... ( -36.10) >DroEre_CAF1 51730 115 + 1 CAGUUGGCCAAAACCGGCCGUGGUCGUAGUGAAGUU----G-GUCUCUCGCACAGGCCAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU .....((((......)))).(((.(((((((..(((----(-((((.......))))))))((....))..)))...((((((((((((....))))).)))))))...)))).)))... ( -32.70) >DroYak_CAF1 50144 116 + 1 CAGUUGGCCAAAACCGGCCGUGGUCGUAGUGAAGUC----GGGGCUCUCGCACAGGCUAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((.....((((....))))..(((((((.----...))).))))...)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... ( -32.10) >DroAna_CAF1 53032 116 + 1 CAGUUGGCCAAAACCGGCCCUGGCC----CGAAGCCUAACCAGGCUCUCGCACAGGCCAACGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((......(((....)))----((((((((....))))).)))....)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... ( -37.10) >consensus CAGUUGGCCAAAACCGGCCGUGGUCGCAGUGAAGUC____GAGGCUCUCGCACAGGCCAGCGCUGUUGCAACAUGUCAAAAUAAAUCAAUUGAUUGAUCUUGUUUUAUUUAUGUCCAAUU ..(((((((.....((((....))))..((((((((......)))).))))...)))))))((....)).(((((..((((((((((((....))))).)))))))...)))))...... (-33.85 = -33.47 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:40 2006