
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,296,970 – 15,297,090 |
| Length | 120 |
| Max. P | 0.721895 |

| Location | 15,296,970 – 15,297,090 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
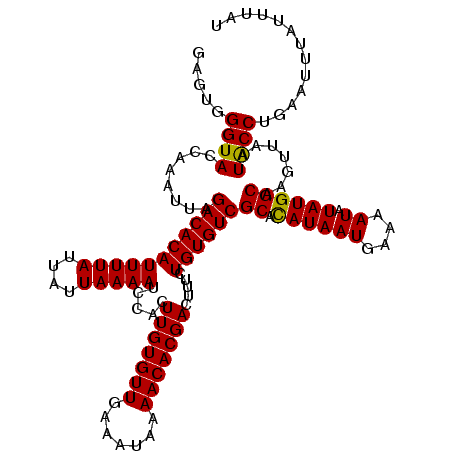
>2L_DroMel_CAF1 15296970 120 + 22407834 GAGUGGGUACCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCACAUAAUGAAAAUAUAUGGCAAGUUAAUGCCGGCAAAUAUUUAU ..((.((((........(((((((((((....))))).....(((((((.......))))))).....))))))((.((((((....)).)))))).......)))).)).......... ( -26.90) >DroVir_CAF1 52988 120 + 1 GAGUGGGUACCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCAUAUAAUGAAAAUAUAUGGCAAGUUAAUACCUGAAUUUAUUUAU ....(((((........(((((((((((....))))).....(((((((.......))))))).....))))))(((((((.(....)))))).)).......)))))............ ( -22.20) >DroGri_CAF1 48192 120 + 1 GAGUGGGUACCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCAUAUAAUGAAAAUAUAUGGCAAGUUAAUACCUGAAUUUAUUUAU ....(((((........(((((((((((....))))).....(((((((.......))))))).....))))))(((((((.(....)))))).)).......)))))............ ( -22.20) >DroYak_CAF1 46483 120 + 1 GAGUGGGUACCAAAUUAGACACAUUUUAUUAAUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCACAUAAUGAAAAUAUAUGGCAAGUUAAUGCCAGAAAAUAUUUAU ..((((....)......(((((((((((....))))).....(((((((.......))))))).....)))))).))).......((((((.(((((......)))))....)))))).. ( -23.40) >DroMoj_CAF1 59695 120 + 1 GAGUGGGUACCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCAUAUAAUGAAAAUAUAUGGCAAGUUAAUACCUGAAUUUAUUUAU ....(((((........(((((((((((....))))).....(((((((.......))))))).....))))))(((((((.(....)))))).)).......)))))............ ( -22.20) >DroAna_CAF1 49518 120 + 1 GAGUGGGUAUCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCCCAUAAUGAGAAUAUAUGGCAAGUUAAUGCCAGAAUUUAUUUAA ..((((((.........(((((((((((....))))).....(((((((.......))))))).....)))))))))))).....(((((..(((((......))))).....))))).. ( -27.60) >consensus GAGUGGGUACCAAAUUAGACACAUUUUAUUAUUAAAAUCCACUUGUGUUGAAAUAAAACACGACUUUCUGUGUCGCACAUAAUGAAAAUAUAUGGCAAGUUAAUACCUGAAUUUAUUUAU .....((((........(((((((((((....))))).....(((((((.......))))))).....))))))((.((((((....)).)))))).......))))............. (-20.68 = -21.43 + 0.75)


| Location | 15,296,970 – 15,297,090 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15296970 120 - 22407834 AUAAAUAUUUGCCGGCAUUAACUUGCCAUAUAUUUUCAUUAUGUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGGUACCCACUC .........((((((((......))))(((((((((.((((((..((((.......))))..)(((((((......))))))).....)))))))))))))).......))))....... ( -23.50) >DroVir_CAF1 52988 120 - 1 AUAAAUAAAUUCAGGUAUUAACUUGCCAUAUAUUUUCAUUAUAUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGGUACCCACUC .............((((((((...((.(((((.......)))))))((((((.(((((((((((.......))))).....))))))..........))))))....))))))))..... ( -22.10) >DroGri_CAF1 48192 120 - 1 AUAAAUAAAUUCAGGUAUUAACUUGCCAUAUAUUUUCAUUAUAUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGGUACCCACUC .............((((((((...((.(((((.......)))))))((((((.(((((((((((.......))))).....))))))..........))))))....))))))))..... ( -22.10) >DroYak_CAF1 46483 120 - 1 AUAAAUAUUUUCUGGCAUUAACUUGCCAUAUAUUUUCAUUAUGUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAUUAAUAAAAUGUGUCUAAUUUGGUACCCACUC ......(((((((((((......)))((((((.......)))))).....)))))))).(((((.......))))).(((((((((((....))))))((..(......)..))))))). ( -24.40) >DroMoj_CAF1 59695 120 - 1 AUAAAUAAAUUCAGGUAUUAACUUGCCAUAUAUUUUCAUUAUAUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGGUACCCACUC .............((((((((...((.(((((.......)))))))((((((.(((((((((((.......))))).....))))))..........))))))....))))))))..... ( -22.10) >DroAna_CAF1 49518 120 - 1 UUAAAUAAAUUCUGGCAUUAACUUGCCAUAUAUUCUCAUUAUGGGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGAUACCCACUC ..(((((((...(((((......)))))................(((((.......)))))..))))))).......(((((((((((....))))))(((((......)))))))))). ( -25.20) >consensus AUAAAUAAAUUCAGGCAUUAACUUGCCAUAUAUUUUCAUUAUAUGCGACACAGAAAGUCGUGUUUUAUUUCAACACAAGUGGAUUUUAAUAAUAAAAUGUGUCUAAUUUGGUACCCACUC .............((((......))))(((((.......)))))..(((.......)))(((((.......))))).(((((((((((....))))))(((((......)))))))))). (-19.92 = -19.45 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:37 2006