
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,283,771 – 15,283,887 |
| Length | 116 |
| Max. P | 0.999124 |

| Location | 15,283,771 – 15,283,887 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.06 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.88 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15283771 116 + 22407834 UUUUUGUCUUCUCUCCUCACGUUGGCGCUAAUGCAAAUGUGAAAAGGCAUUUUGUGAGCCAAAAUGAGGU----UGUUGUCGUGCCACUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((((((.......)))----)(((((((......)))))))..........)))))))))))))). ( -32.30) >DroPse_CAF1 32493 120 + 1 UUUUUGUCUUCUCUCUUCACGUUGGCGCUAAUGCAAAUGUGGAAAGGCAUUUUGUGAGGCAAAAUGAGGUUGGUUGUUGUCGUGCCGCUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((.((((..(..(.(.....).)..)..)))).((((........))))..)))))))))))))). ( -32.10) >DroSec_CAF1 31063 116 + 1 UUUUUGUCUUCUCUCCUCACGUUGGCGCUAAUGCAAAUGUGAAAAGGCAUUUUGUGAGCCAAAAUGAGGU----UGUUGUCGUGCCACUGACAAUCUAAUUAGAAUACGAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((((((.......)))----)(((((((......)))))))..........)))))))))))))). ( -32.00) >DroYak_CAF1 32701 116 + 1 UUUUUGUCUUCUCUCCUCACGUUGGCGCUAAUGCAAAUGUGAAAAGGCAUUUUGUGAGCCAAAAUGAGGU----UGUUGUCGUGCCACUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((((((.......)))----)(((((((......)))))))..........)))))))))))))). ( -32.30) >DroAna_CAF1 32831 116 + 1 UUUUUGUCUUCUCUCCUCACGUUGGCGCUAAUGCAAAUGUGAAAAGGCAUUUUGUGAGCCAAAAUGAGGU----UGUUGUCGUGCCGCUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((((((.......)))----)(((((((......)))))))..........)))))))))))))). ( -32.30) >DroPer_CAF1 32868 120 + 1 UUUUUGUCUUCUCUCUUCACGUUGGCGCUAAUGCAAAUGUGGAAAGGCAUUUUGUGAGGCAAAAUGAGGUUGGUUGUUGUCGUGCCGCUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((.((((..(..(.(.....).)..)..)))).((((........))))..)))))))))))))). ( -32.10) >consensus UUUUUGUCUUCUCUCCUCACGUUGGCGCUAAUGCAAAUGUGAAAAGGCAUUUUGUGAGCCAAAAUGAGGU____UGUUGUCGUGCCACUGACAAUCUAAUUAGAAUACAAAAUGCUUUUA ................(((((((.(((....))).)))))))((((((((((((((....................((((((......))))))(((....))).)))))))))))))). (-28.88 = -28.52 + -0.36)


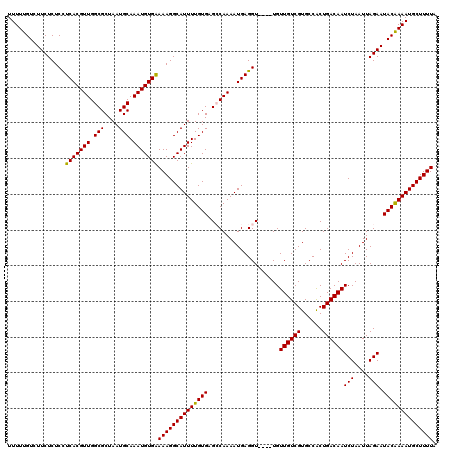
| Location | 15,283,771 – 15,283,887 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.06 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -25.53 |
| Energy contribution | -26.37 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15283771 116 - 22407834 UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGUGGCACGACAACA----ACCUCAUUUUGGCUCACAAAAUGCCUUUUCACAUUUGCAUUAGCGCCAACGUGAGGAGAGAAGACAAAAA ........((((((.((((.......(((((........)))))..----.((((((.(((((.....(((((........)))))((....))))))).))))))..)))).)))))). ( -30.90) >DroPse_CAF1 32493 120 - 1 UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGCGGCACGACAACAACCAACCUCAUUUUGCCUCACAAAAUGCCUUUCCACAUUUGCAUUAGCGCCAACGUGAAGAGAGAAGACAAAAA .....(((((((((..(((....)))(((((........))))).....................)))))))))((((((((.((.((......)).)).)))..))))).......... ( -21.50) >DroSec_CAF1 31063 116 - 1 UAAAAGCAUUUCGUAUUCUAAUUAGAUUGUCAGUGGCACGACAACA----ACCUCAUUUUGGCUCACAAAAUGCCUUUUCACAUUUGCAUUAGCGCCAACGUGAGGAGAGAAGACAAAAA ............((.((((.......(((((........)))))..----.((((((.(((((.....(((((........)))))((....))))))).))))))..)))).))..... ( -27.50) >DroYak_CAF1 32701 116 - 1 UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGUGGCACGACAACA----ACCUCAUUUUGGCUCACAAAAUGCCUUUUCACAUUUGCAUUAGCGCCAACGUGAGGAGAGAAGACAAAAA ........((((((.((((.......(((((........)))))..----.((((((.(((((.....(((((........)))))((....))))))).))))))..)))).)))))). ( -30.90) >DroAna_CAF1 32831 116 - 1 UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGCGGCACGACAACA----ACCUCAUUUUGGCUCACAAAAUGCCUUUUCACAUUUGCAUUAGCGCCAACGUGAGGAGAGAAGACAAAAA ........((((((.((((.......(((((........)))))..----.((((((.(((((.....(((((........)))))((....))))))).))))))..)))).)))))). ( -30.90) >DroPer_CAF1 32868 120 - 1 UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGCGGCACGACAACAACCAACCUCAUUUUGCCUCACAAAAUGCCUUUCCACAUUUGCAUUAGCGCCAACGUGAAGAGAGAAGACAAAAA .....(((((((((..(((....)))(((((........))))).....................)))))))))((((((((.((.((......)).)).)))..))))).......... ( -21.50) >consensus UAAAAGCAUUUUGUAUUCUAAUUAGAUUGUCAGCGGCACGACAACA____ACCUCAUUUUGGCUCACAAAAUGCCUUUUCACAUUUGCAUUAGCGCCAACGUGAGGAGAGAAGACAAAAA ........((((((.((((.......(((((........))))).......((((((.(((((.....(((((........)))))((....))))))).))))))..)))).)))))). (-25.53 = -26.37 + 0.83)

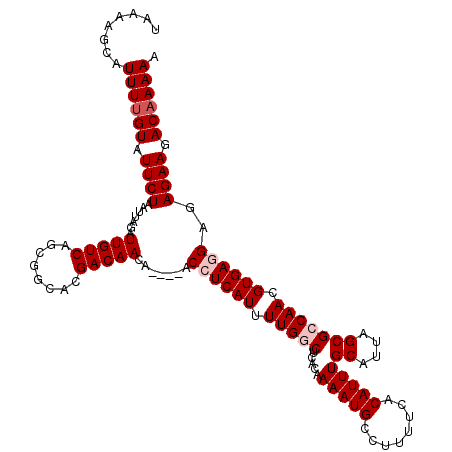
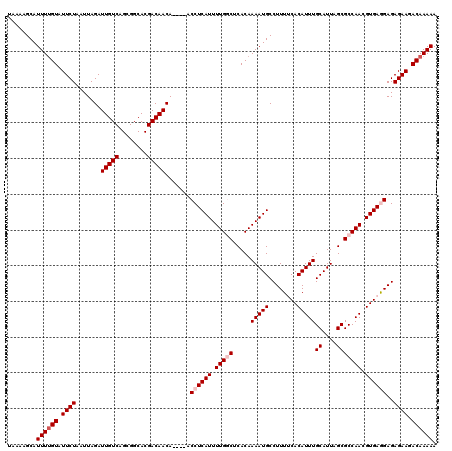
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:31 2006