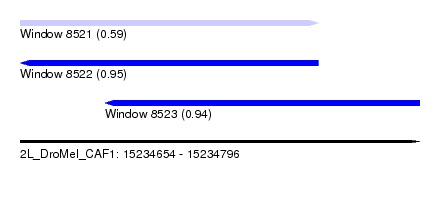
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,234,654 – 15,234,796 |
| Length | 142 |
| Max. P | 0.947005 |

| Location | 15,234,654 – 15,234,760 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15234654 106 + 22407834 -------UUGGUUUUACCU-AGCUUUUGCAUUUC-GU-UU--UCUGCGUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUCGGCAACGGCAGCC -------..((.....)).-.((....)).....-..-..--.((((..(((((((((((....))))))........(..(((((((.......)))))))..))))))..)))).. ( -23.60) >DroVir_CAF1 249414 101 + 1 -------UUGGCGUUC-------GUUUGUGUUCA-GCGUU--UCUGCGUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUUGGCAGCGGCAAUG -------(((.(((..-------..........(-((((.--...)))))((((((((((....))))))...........(((((((.......)))))))...))))))).))).. ( -22.20) >DroPse_CAF1 253972 102 + 1 -------UUAGUUUUGGGU-UCGUUUUGAGUUUC-GU-UU--UCUGCGUUUGCCGCUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUCGGCAACGGC---- -------............-((((((((((((..-..-..--...((((((((.((.......))))))))))..........(((((.......)))))))))))).))))).---- ( -20.70) >DroGri_CAF1 221444 91 + 1 -------U-----------------UUGUGUUCA-GCAUU--GCUGCAUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUUGGCAACGGCAAUG -------.-----------------.........-.((((--((((...(((((((((((....))))))...........(((((((.......)))))))...))))))))))))) ( -23.70) >DroWil_CAF1 274323 117 + 1 CUUAUGUUUAGUUUUAGUUUUCAUUUUGCGUUGCCGU-UGCCUGUGCGUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUCGGCAAUGGCAACG ............................(((((((((-((((((..((.((((....)))).))..))..........(..(((((((.......)))))))..)))))))))))))) ( -36.80) >DroMoj_CAF1 276136 101 + 1 -------UUGGCGUUC-------GUUUGUGCUCA-GCGCU--GCUGCGUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUUGGCAACGGCAAUG -------(((.(((((-------(((((..(...-((((.--...))))((((....))))..)..)))))).(((..((..((((((.......))))))))..))))))).))).. ( -26.60) >consensus _______UUGGUUUUA_______UUUUGUGUUCA_GC_UU__UCUGCGUUUGCCUUUGCAAUUGUGCAAAUGUGUUUUGUUUUGAUAAAUUAGGCUUGUCAACUCGGCAACGGCAAUG ............................................(((..(((((((((((....))))))...........(((((((.......)))))))...)))))..)))... (-16.95 = -17.32 + 0.36)

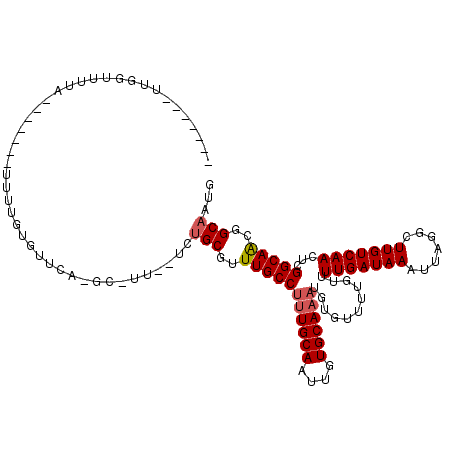
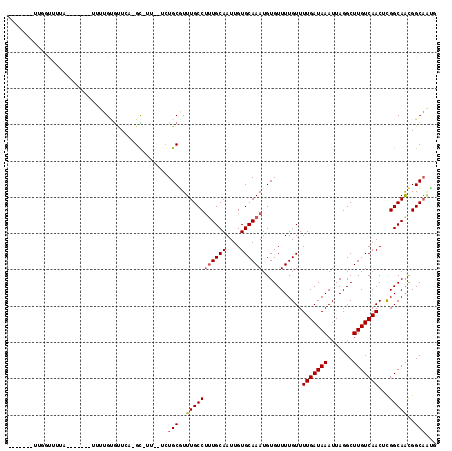
| Location | 15,234,654 – 15,234,760 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -14.54 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15234654 106 - 22407834 GGCUGCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA--AA-AC-GAAAUGCAAAAGCU-AGGUAAAACCAA------- ..((((..((((((..((((.(((.....))).))))..)........((((((....)))))))))))..)))).--..-..-.....((....)).-.((.....))..------- ( -21.30) >DroVir_CAF1 249414 101 - 1 CAUUGCCGCUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA--AACGC-UGAACACAAAC-------GAACGCCAA------- ..((((...((((...((((.(((.....))).))))...........((((((....))))))))))...)))).--.....-...........-------.........------- ( -14.90) >DroPse_CAF1 253972 102 - 1 ----GCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAGCGGCAAACGCAGA--AA-AC-GAAACUCAAAACGA-ACCCAAAACUAA------- ----..((((...(((((................................((((....))))(((.....)))...--..-..-..)))))..)))).-............------- ( -15.70) >DroGri_CAF1 221444 91 - 1 CAUUGCCGUUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAAUGCAGC--AAUGC-UGAACACAA-----------------A------- ((((((.((((((...((((.(((.....))).))))...........((((((....))))))))))...)).))--)))).-.........-----------------.------- ( -18.80) >DroWil_CAF1 274323 117 - 1 CGUUGCCAUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCACAGGCA-ACGGCAACGCAAAAUGAAAACUAAAACUAAACAUAAG (((((((.(((((..(((.....((((..(((........)))......(((((....))))))))).))).....))))-).)))))))............................ ( -26.70) >DroMoj_CAF1 276136 101 - 1 CAUUGCCGUUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGC--AGCGC-UGAGCACAAAC-------GAACGCCAA------- ...((((((((((...((((.(((.....))).))))...........((((((....))))))))).))))((((--...))-)).))).....-------.........------- ( -18.60) >consensus CAUUGCCGUUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA__AA_AC_GGAACACAAAA_______GAAAACCAA_______ ..((((..(((((((((((.......))))))................((((((....)))))))))))..))))........................................... (-14.54 = -14.90 + 0.36)



| Location | 15,234,684 – 15,234,796 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -15.03 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15234684 112 - 22407834 CUCCUUCGCCCACCGCACCACCCAUUUUGGCGCU----CUG--GCUGCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA .........(((..((.(((.......))).)).----.))--)((((..((((((..((((.(((.....))).))))..)........((((((....)))))))))))..)))). ( -27.10) >DroVir_CAF1 249439 84 - 1 ----------------------------CUCGCU----CCC--AUUGCCGCUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA ----------------------------......----...--.((((...((((...((((.(((.....))).))))...........((((((....))))))))))...)))). ( -14.90) >DroPse_CAF1 254002 101 - 1 CCUCUUC-----AUGCCCCACCCAUUGUGGCGCC------------GCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAGCGGCAAACGCAGA ..(((.(-----..((.((((.....)))).)).------------((((((((.((((((.......)))))).......((((.....)))).......))))))))....).))) ( -22.60) >DroEre_CAF1 184157 111 - 1 -CUCUGCGCCCACCGCACCACCCAUUUUGGCGCU----UGG--GCUGCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA -.((((((((((..((.(((.......))).)).----)))--))(((((((......((((.(((.....))).)))).....)))...((((((....))))))))))...))))) ( -34.70) >DroMoj_CAF1 276161 85 - 1 ---------------------------UCUCGUU----UCC--AUUGCCGUUGCCAAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGC ---------------------------.......----...--.((((..(((((...((((.(((.....))).))))...........((((((....)))))))))))..)))). ( -16.20) >DroAna_CAF1 190841 108 - 1 CUUCUU----------UCCACCCAUUUUGGCGCUAGCUCCGGCGCUGGCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA ......----------............((((((......)))))).(((((((((..((((.(((.....))).))))..)........((((((....))))))))).)))))... ( -27.00) >consensus __UCUU___________CCACCCAUUUUGGCGCU____CCG__GCUGCCGUUGCCGAGUUGACAAGCCUAAUUUAUCAAAACAAAACACAUUUGCACAAUUGCAAAGGCAAACGCAGA ............................................((((..(((((((((((.......))))))................((((((....)))))))))))..)))). (-15.03 = -15.33 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:25 2006