
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,232,787 – 15,232,905 |
| Length | 118 |
| Max. P | 0.999703 |

| Location | 15,232,787 – 15,232,905 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15232787 118 + 22407834 AGCGAAAAAUUUCCACUUGCCACCCGAUAAGGAUGGCAAAACUUUGUUACUUUGGCGAUGGC-GACAACUUUGCCAACUAACAAACUAUUAUAUGCAAACAAUAAUUGCACUUUAUUUG .((((...........((((((.((.....)).))))))...(((((((..(((((((((..-..))...))))))).)))))))....................)))).......... ( -28.30) >DroSec_CAF1 177008 117 + 1 AGCAAAAAAUUUCCACUUGCCACCCGAUAAGGAUGGCAAAACUUUGUUACUUUGGCGAUGGC-GACAACUUUGCCAACUAGCAAACUAUUAUAUGCAAACAAUAAUUGCACUUU-UGUG .((((((.........((((((.((.....)).))))))...(((((((..(((((((((..-..))...))))))).)))))))........(((((.......))))).)))-))). ( -30.70) >DroSim_CAF1 183717 118 + 1 AGCAAAAAAUUUCCACUUGCCACCCGAUAAGGAUGGCAAAACUUUGUUACUUUGGCGAUGGC-GACAACUUUGCCAACGAGCAAACUAUUAUAUGCAAACAAUAAUUGCACUUUAUAUG .((((...........((((((.((.....)).))))))...((((((.(.(((((((((..-..))...))))))).)))))))....................)))).......... ( -28.50) >DroEre_CAF1 182410 118 + 1 AGCAAAAAAUUUCCAC-UGCCACCCGAUAAGGAUGGCAAAACUUUGUUUCUUUGGCGCUGGGCGACAACUUUGCCAACUAGCAAACUGCUAUAUGCAAGAAAUAAUUGCACUUUACAUG .((((...........-(((((.((.....)).))))).....((((((((...(((...(((((.....)))))...((((.....))))..))).)))))))))))).......... ( -31.40) >DroYak_CAF1 193728 118 + 1 AGCAAAAAAUGUCCACUUGCCACCCGACAAGGAUGGCAAAACUUUGUUACUUUGGCCAUGGG-GACAACUUUGCCAACUAACAAACUAUUAUAUGCAAACAAUAAUUGCACUUAAUAUG ................((((((.((.....)).))))))...(((((((..(((((..((..-..)).....))))).))))))).(((((..(((((.......)))))..))))).. ( -29.20) >consensus AGCAAAAAAUUUCCACUUGCCACCCGAUAAGGAUGGCAAAACUUUGUUACUUUGGCGAUGGC_GACAACUUUGCCAACUAGCAAACUAUUAUAUGCAAACAAUAAUUGCACUUUAUAUG .((((...........((((((.((.....)).))))))...((((((...(((((((.(........).)))))))..))))))....................)))).......... (-25.74 = -25.94 + 0.20)

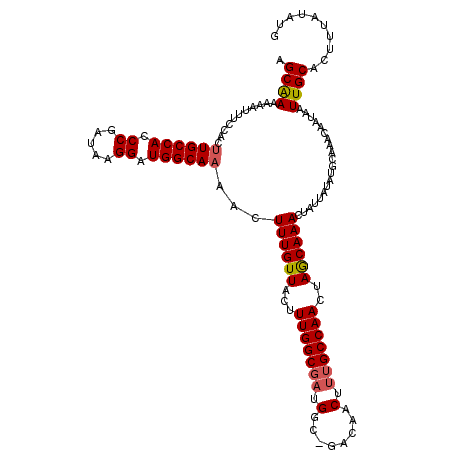
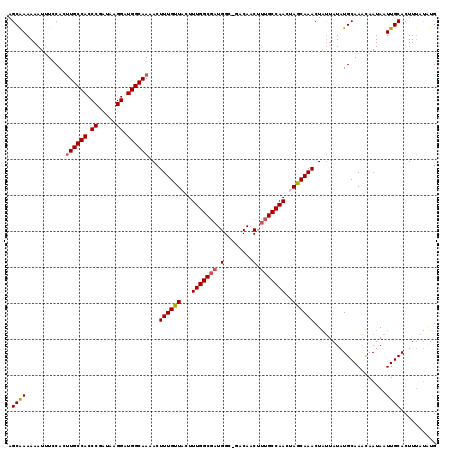
| Location | 15,232,787 – 15,232,905 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -26.05 |
| Energy contribution | -25.69 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15232787 118 - 22407834 CAAAUAAAGUGCAAUUAUUGUUUGCAUAUAAUAGUUUGUUAGUUGGCAAAGUUGUC-GCCAUCGCCAAAGUAACAAAGUUUUGCCAUCCUUAUCGGGUGGCAAGUGGAAAUUUUUCGCU ........((((((.......)))))).......(((((((.(((((.........-......)))))..)))))))...(((((((((.....)))))))))(((((.....))))). ( -33.46) >DroSec_CAF1 177008 117 - 1 CACA-AAAGUGCAAUUAUUGUUUGCAUAUAAUAGUUUGCUAGUUGGCAAAGUUGUC-GCCAUCGCCAAAGUAACAAAGUUUUGCCAUCCUUAUCGGGUGGCAAGUGGAAAUUUUUUGCU ..((-(....(((((((((((......))))))).))))...)))((((((..((.-.((((.((....)).........(((((((((.....)))))))))))))..)).)))))). ( -31.20) >DroSim_CAF1 183717 118 - 1 CAUAUAAAGUGCAAUUAUUGUUUGCAUAUAAUAGUUUGCUCGUUGGCAAAGUUGUC-GCCAUCGCCAAAGUAACAAAGUUUUGCCAUCCUUAUCGGGUGGCAAGUGGAAAUUUUUUGCU ..........((((.....((((.(((....((.((((..((.((((.........-)))).)).)))).))........(((((((((.....)))))))))))).))))...)))). ( -31.50) >DroEre_CAF1 182410 118 - 1 CAUGUAAAGUGCAAUUAUUUCUUGCAUAUAGCAGUUUGCUAGUUGGCAAAGUUGUCGCCCAGCGCCAAAGAAACAAAGUUUUGCCAUCCUUAUCGGGUGGCA-GUGGAAAUUUUUUGCU ..........((((..(((((((((.....)))((((.((..(((((...((((.....)))))))))))))))......(((((((((.....))))))))-).))))))...)))). ( -33.10) >DroYak_CAF1 193728 118 - 1 CAUAUUAAGUGCAAUUAUUGUUUGCAUAUAAUAGUUUGUUAGUUGGCAAAGUUGUC-CCCAUGGCCAAAGUAACAAAGUUUUGCCAUCCUUGUCGGGUGGCAAGUGGACAUUUUUUGCU ..........((((..(.((((..(.........(((((((.(((((.....((..-..))..)))))..)))))))...(((((((((.....))))))))))..)))).)..)))). ( -37.80) >consensus CAUAUAAAGUGCAAUUAUUGUUUGCAUAUAAUAGUUUGCUAGUUGGCAAAGUUGUC_GCCAUCGCCAAAGUAACAAAGUUUUGCCAUCCUUAUCGGGUGGCAAGUGGAAAUUUUUUGCU ........((((((.......))))))(((((..((((((....)))))))))))........((.((((.........((((((((((.....)))))))))).......)))).)). (-26.05 = -25.69 + -0.36)

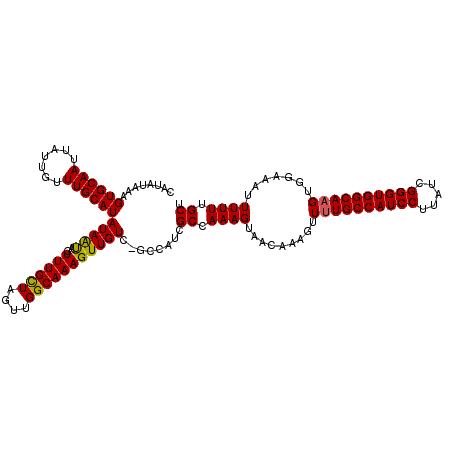
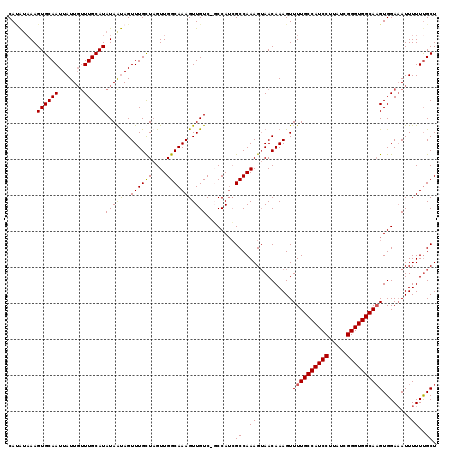
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:23 2006