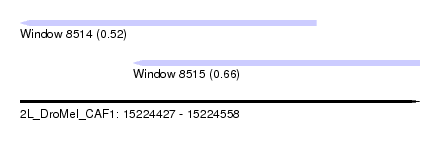
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,224,427 – 15,224,558 |
| Length | 131 |
| Max. P | 0.658384 |

| Location | 15,224,427 – 15,224,524 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.89 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -7.86 |
| Energy contribution | -7.95 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.31 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15224427 97 - 22407834 ------GUAUAAUAUAUAACGCUCCUAU-----GAUGGCGUUCACUUCAUCAUAAGCUGGUGACAGUGU--GU--UGCAAGCUGGUGAUAAAUAUCACAC-UCAUAAUAAUCU ------..........((((((...(((-----(((((.(....).)))))))).((((....))))))--))--)).......(((((....)))))..-............ ( -23.50) >DroSec_CAF1 168651 77 - 1 -------------------------U--------AUGGCGUUCACUUCAUCAUAAGCUGGUGACAGUGU--GUGGUGCAAGCUGGUGAUAAAUAUCACAC-UCAUAAUAAUCU -------------------------(--------((((.((.((((((((((((.((((....)))).)--)))))).))).))(((((....)))))))-)))))....... ( -24.30) >DroSim_CAF1 171968 96 - 1 ------GUGUAAUGUAUAACGCUCGU--------AUGGCGUUCACUUCAUCAUAAGCUGGUGACAUUGU--GUGGUGCAAGCUGGUGAUAAAUAUCACAC-UCAUAAUAAUCU ------((((.((((...(((((...--------..)))))(((((.(((((((...((....))...)--))))))......)))))...)))).))))-............ ( -23.30) >DroEre_CAF1 174096 85 - 1 --------------UAUAACGCUCGU--------AUGGCGU-CACUUCAUCAUAAGCUGGUGACAGUGU--GU--UGCAAGCUGGUGAUAAAUAUUACAC-UCGUGAUAAUCU --------------....(((((...--------..)))))-......(((((.((((.(..((.....--))--..).)))).)))))...((((((..-..)))))).... ( -22.40) >DroYak_CAF1 184918 100 - 1 AUGUAUGUAUAAUAUAUAACGCUUAU--------AUGGCGUUCACUUCAUCAUGAGCUGGUGACAGUGU--GG--UGCAAGCUGGUGAUAAAUAUCACAC-UCAUGUUAAUCU ((((((.....))))))((((((...--------..))))))(((((((((((..((((....))))))--))--)).))).))(((((....)))))..-............ ( -27.00) >DroAna_CAF1 180287 104 - 1 ------UGAUACUACACACC-CACCCAUCCCGGGGUG-GGUGCACUUCAUCAUGAACAGGUGGCCCUGCUGGCGGUGGAUGAUGUUGAUAAAUUCC-CAUAAAGUAAUACCCU ------...((((...((((-(((((......)))))-))))..(..((((((..((.(.((((...)))).).))..))))))..).........-.....))))....... ( -31.80) >consensus ______GUAUAAUAUAUAACGCUCCU________AUGGCGUUCACUUCAUCAUAAGCUGGUGACAGUGU__GU__UGCAAGCUGGUGAUAAAUAUCACAC_UCAUAAUAAUCU ................................................(((((.(((((....)..(((.......))))))).)))))........................ ( -7.86 = -7.95 + 0.09)


| Location | 15,224,464 – 15,224,558 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.26 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -12.46 |
| Energy contribution | -13.82 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15224464 94 - 22407834 GCUGGAGUGGUUUAACAUAUUCCGCUUUACAUGU----------GUAUAAUAUAUAACGCUCCUAU-----GAUGGCGUUCACUUCAUCAUAAGCUGGUGACAGUGU--GU-- ..((((((((..((...))..))))))))..(((----------(((...))))))((((...(((-----(((((.(....).)))))))).((((....))))))--))-- ( -24.50) >DroSim_CAF1 172005 93 - 1 GCUGGAGUGGUUUAACAUAUUCCGCUUUACAUGU----------GUGUAAUGUAUAACGCUCGU--------AUGGCGUUCACUUCAUCAUAAGCUGGUGACAUUGU--GUGG ..((((((((............((((.(((..((----------((..........))))..))--------).)))).)))))))).((((...((....))...)--))). ( -22.62) >DroEre_CAF1 174133 83 - 1 GCUGGAGUGGUUUACCAUACUCCGCUUUACAUAUG-----------------UAUAACGCUCGU--------AUGGCGU-CACUUCAUCAUAAGCUGGUGACAGUGU--GU-- (.((((((((....((((((..((...(((....)-----------------))...))...))--------))))..)-))))))).)....((((....))))..--..-- ( -24.50) >DroYak_CAF1 184955 101 - 1 GCUGGAGUGGUUUAACAUAUUCCGCUUUACAUAUAUAUAUGUAUGUAUAAUAUAUAACGCUUAU--------AUGGCGUUCACUUCAUCAUGAGCUGGUGACAGUGU--GG-- ..(((((((.......(((((..((..((((((....)))))).))..)))))..((((((...--------..))))))))))))).(((..((((....))))))--).-- ( -27.50) >DroAna_CAF1 180324 101 - 1 CAUGAAGCUGUUUAACAUAAUCUGCUUUACGUGA----------UGAUACUACACACC-CACCCAUCCCGGGGUG-GGUGCACUUCAUCAUGAACAGGUGGCCCUGCUGGCGG ..((((((.(((......)))..))))))(((((----------(((.......((((-(((((......)))))-))))....))))))))..((((....))))....... ( -36.60) >consensus GCUGGAGUGGUUUAACAUAUUCCGCUUUACAUGU__________GUAUAAUAUAUAACGCUCCU________AUGGCGUUCACUUCAUCAUAAGCUGGUGACAGUGU__GU__ ..((((((((...........))))))))..........................(((((.(((........))))))))((((((((((.....)))))).))))....... (-12.46 = -13.82 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:18 2006