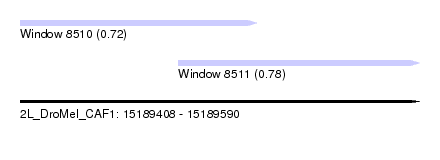
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,189,408 – 15,189,590 |
| Length | 182 |
| Max. P | 0.784545 |

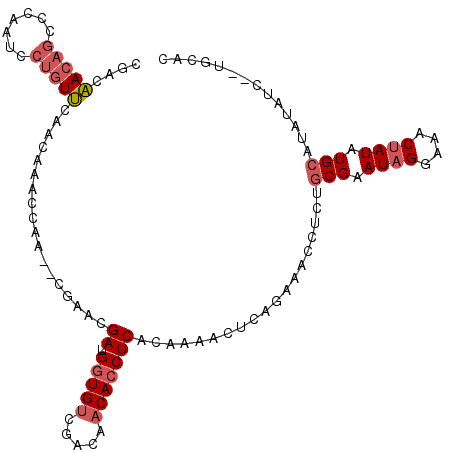
| Location | 15,189,408 – 15,189,516 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -13.14 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15189408 108 + 22407834 CGACAACAGCCCAAUCCUGUUCAGCAAACCAA--CGAACGAUGGUGUCGACAACACCUCACAAAUCUCAGAAACCCCUGCGAAUAGGAAACUAUAUGCAUAUAUC--UGCUU (((((((((.......)))))......((((.--(....).))))))))..................((((......((((.((((....)))).))))....))--))... ( -22.30) >DroSec_CAF1 134344 108 + 1 CGACAACAGCCCAAUCCUGUUCAACAAACCAA--CGAACGAUGGUGUCGACAACACCUCACAAGACUCACAAACCUCUGCGAAUAGGAAACUAUAUGCAUAUAUC--UGCAC (((((((((.......)))))......((((.--(....).))))))))............................((((.((((....)))).))))......--..... ( -20.30) >DroSim_CAF1 137679 108 + 1 CGACAACAGCCCAAUCCUGUUCAACAAACCAA--CGAACGAUGGUGUCGACAACACCUCACAAAACUCACAAACCUCUGCGAAUAGGAAACUAUAUGCAUAUAUC--UGCAC (((((((((.......)))))......((((.--(....).))))))))............................((((.((((....)))).))))......--..... ( -20.30) >DroEre_CAF1 137336 97 + 1 CGACAACAGCCCAAUCCUGUUCAGCAAACCAG--CGAACGAUGGUGGCGACAACACCUCACAAGACUCAGAGUCC-CGGCGAAUUGGAAACUAUAUG------------CAC .(((..............((((.((......)--)))))((.((((.......))))))............))).-..(((.((.(....).)).))------------).. ( -19.70) >DroYak_CAF1 149879 108 + 1 CGACAACAGCCCAAUCCUGUUCAACAAACCAA--CGAAGGACGGUGUCGACAACACCUCACAGAACUCAGAAACCUCUGCGAAUAGGAAACUAUAUGCAUAUAUC--UGCAC (((((((((.......)))))......(((..--(....)..)))))))...........((((....(((....)))(((.((((....)))).))).....))--))... ( -23.60) >DroPer_CAF1 171410 98 + 1 CGAGGACAAUCCAAAACCAUCGA--AAACCAAACCAAACGACGAUGUCGACAACAACUCACAAA------------CAGCGAAUAGGAAUCUAUAUGCAUAUAUCCUUGCAA ((((((...........(((((.--................)))))..................------------..(((.((((....)))).))).....))))))... ( -14.63) >consensus CGACAACAGCCCAAUCCUGUUCAACAAACCAA__CGAACGAUGGUGUCGACAACACCUCACAAAACUCAGAAACCUCUGCGAAUAGGAAACUAUAUGCAUAUAUC__UGCAC ....(((((.......)))))..................((.(((((.....)))))))...................(((.((((....)))).))).............. (-13.14 = -14.03 + 0.89)

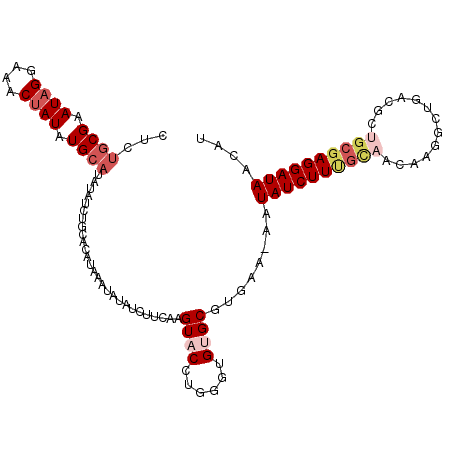
| Location | 15,189,480 – 15,189,590 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -15.70 |
| Energy contribution | -17.46 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15189480 110 + 22407834 CCCUGCGAAUAGGAAACUAUAUGCAUAUAUCUGCUUAUAAAUAUAUCUUGAAGUACCUGGGUGUGCGUGAA-AAUAUCUUUGUAACAAGGCUGACGCAGCGAGGAUAACAU ...((((.((((....)))).))))..(((((..............((((...(((..((((((.......-.))))))..))).))))((((...))))..))))).... ( -26.40) >DroSec_CAF1 134416 111 + 1 CUCUGCGAAUAGGAAACUAUAUGCAUAUAUCUGCACAUAAAUAUAUCUUCAAGUACCUGGGUGCGCGUGAAAAAUAUCUUUGCAACAAUGCUGCCGCUGCGAGGAUAACAU ...((((.((((....)))).))))......................((((.((((....))))...))))...((((((((((.....((....)))))))))))).... ( -28.30) >DroSim_CAF1 137751 111 + 1 CUCUGCGAAUAGGAAACUAUAUGCAUAUAUCUGCACAUAAAUAUAUCUUCAAGUACCUGGGUGCGCGUGAAAAAUAUCUUCGCAACAAUGCUGCCGCUGCGAGGAUAACAU ...((((.((((....)))).))))......................((((.((((....))))...))))...((((((((((.....((....)))))))))))).... ( -30.40) >DroEre_CAF1 137408 85 + 1 C-CGGCGAAUUGGAAACUAUAUG----------CACAUAAAUAUUUUUCCAAGUACCCGGCUGUGCGUAAU-AAUAUCUUUGCAACA--------------AGGAUAACAU (-(((....(((((((..((((.----------.......)))).)))))))....))))...........-..((((((((...))--------------)))))).... ( -19.90) >DroYak_CAF1 149951 109 + 1 CUCUGCGAAUAGGAAACUAUAUGCAUAUAUCUGCACAUAAAUAUUUCUUUAGGUACCUGUGUGUGCGUAAA-AAUAUCUUUGCAACA-GACUGACGCUGCGAGGAUAACAU ...((((.((((....)))).)))).......(((((((......((....))......))))))).....-..((((((((((.(.-.......).)))))))))).... ( -30.00) >consensus CUCUGCGAAUAGGAAACUAUAUGCAUAUAUCUGCACAUAAAUAUAUCUUCAAGUACCUGGGUGUGCGUGAA_AAUAUCUUUGCAACAAGGCUGACGCUGCGAGGAUAACAU ...((((.((((....)))).))))...........................((((......))))........((((((((((.............)))))))))).... (-15.70 = -17.46 + 1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:14 2006