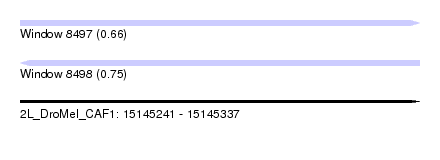
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,145,241 – 15,145,337 |
| Length | 96 |
| Max. P | 0.748515 |

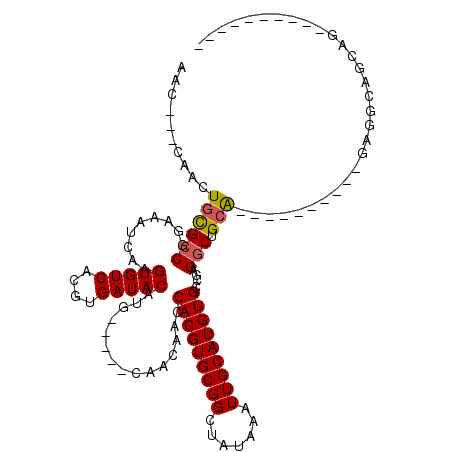
| Location | 15,145,241 – 15,145,337 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15145241 96 + 22407834 ------GCACUCACUUCCUC----------UGCAGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUG-----CAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUG---GUU ------..........((.(----------(((.(((.....(((((((....(((((((....)))))))))-----)))))((((.(.....).))))....)))))))..)---).. ( -28.20) >DroVir_CAF1 121309 100 + 1 ----------CUGCUGUUGC----------UGCUGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUGCAUUGCAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUCCCAGUU ----------..((((..((----------(((.(((.....(((((((((..(((((((....)))))))...)))))))))((((.(.....).))))....))))))))...)))). ( -35.70) >DroPse_CAF1 109769 102 + 1 CUGGUUGCUGCUGCUGCUGC----------UGCAGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUG-----CAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUG---GUU ((..((((.((.(((((...----------.)))))..(((.(((((((....(((((((....)))))))))-----)))))((((.(.....).))))..)))))))))..)---).. ( -35.40) >DroWil_CAF1 155159 92 + 1 ----------UGCCUGCCUG----------CUCAGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUG-----CAUGUAUCACGUGACACUUGAUUUCCGGCACAGUUG---GCU ----------.....(((.(----------((..(((.....(((((((....(((((((....)))))))))-----)))))((((.(.....).))))....)))..))).)---)). ( -28.00) >DroMoj_CAF1 128431 90 + 1 ------------------------------UGCUGGCUUGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUGCAUUGCAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUGCCAGCU ------------------------------.((((((...(((((((((((..(((((((....)))))))...)))))))))((((.(.....).)))).......))....)))))). ( -34.60) >DroAna_CAF1 96605 106 + 1 ------ACAUCCACUUCCUCUUGUUGCUGCUGCAGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUG-----CAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUG---GUU ------....................(.(((((.(((.....(((((((....(((((((....)))))))))-----)))))((((.(.....).))))....)))))))).)---... ( -28.30) >consensus __________CCACUGCCUC__________UGCAGCCUCGGCACAUGCAAUUUAUAGCCGCACGUGGUUGUUG_____CAUGUAUCACGUGACACUUGAUUUCCGGCGCAGUUG___GUU ..............................(((.(((.((((..............))))((((((((...((.....))...)))))))).............)))))).......... (-19.44 = -19.97 + 0.53)

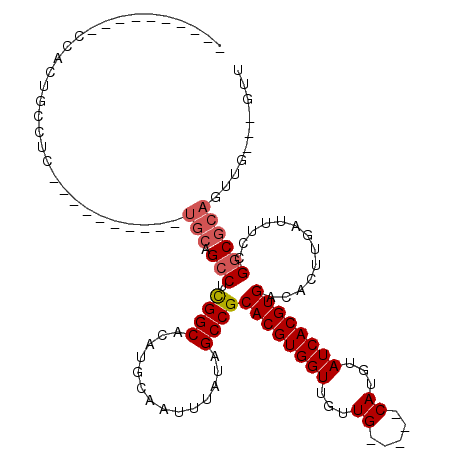
| Location | 15,145,241 – 15,145,337 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15145241 96 - 22407834 AAC---CAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUG-----CAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCUGCA----------GAGGAAGUGAGUGC------ ..(---(..(((((((.........(((((....)))))...-----.......(((((((((.......)))))))))....))).)))----------).))..........------ ( -27.10) >DroVir_CAF1 121309 100 - 1 AACUGGGAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUGCAAUGCAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCAGCA----------GCAACAGCAG---------- ..(((....(((((((((.......(((((....))))).(((...)))....))((((((((.......)))))))).....))).)))----------)...)))...---------- ( -28.60) >DroPse_CAF1 109769 102 - 1 AAC---CAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUG-----CAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCUGCA----------GCAGCAGCAGCAGCAACCAG ...---...((((..(((.......(((((....)))))...-----.......(((((((((.......))))))))))))..(((((.----------...))))).))))....... ( -32.70) >DroWil_CAF1 155159 92 - 1 AGC---CAACUGUGCCGGAAAUCAAGUGUCACGUGAUACAUG-----CAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCUGAG----------CAGGCAGGCA---------- .((---(..((((..(((...((..(((((....)))))...-----.......(((((((((.......)))))))))..))..))).)----------)))...))).---------- ( -29.70) >DroMoj_CAF1 128431 90 - 1 AGCUGGCAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUGCAAUGCAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCAAGCCAGCA------------------------------ .((((((....((((..(....)..))))....((.((((((((((((..((......))..)).....)))))))))).)).)))))).------------------------------ ( -30.10) >DroAna_CAF1 96605 106 - 1 AAC---CAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUG-----CAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCUGCAGCAGCAACAAGAGGAAGUGGAUGU------ ..(---((.((...((((.......(((((....)))))...-----.......(((((((((.......)))))))))))(..((((...))))..)....)).)))))....------ ( -27.30) >consensus AAC___CAACUGCGCCGGAAAUCAAGUGUCACGUGAUACAUG_____CAACAACCACGUGCGGCUAUAAAUUGCAUGUGCCGAGGCUGCA__________GAGGCAGCAG__________ ..........((((((.........(((((....)))))...............(((((((((.......)))))))))....))).))).............................. (-20.08 = -20.13 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:01 2006