
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,122,041 – 15,122,161 |
| Length | 120 |
| Max. P | 0.988736 |

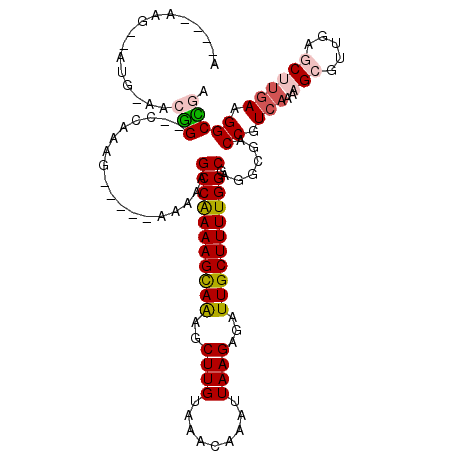
| Location | 15,122,041 – 15,122,144 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15122041 103 + 22407834 A----AAG--AUG-AACGG--CCAAAG-----GAAAAGCCGAAAGCAGAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGAGCUUGAAGGCCGA .----...--...-..(((--((....-----......((((((((((..((((.........))))...)))))))))).(((((...(((.....)))....)))))..))))). ( -33.00) >DroVir_CAF1 93393 104 + 1 ACGGG-------GAAAGAGGGCGGACAGA-----AUGGCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGCCAGUCGGCCCUCAAAAGCGUUGAAC-UGAAGGCUAA .....-------....((((((.(((...-----.(((((((((((((..((((.........))))...)))))))))))))))).))))))...(((.((....-..)).))).. ( -43.70) >DroPse_CAF1 83954 106 + 1 A----AAG--ACG-AACGG--CCAAAG--AAGAAAAAGCCGAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGGGCUUGAAGGCCGA .----...--...-..(((--((....--.........((((((((((..((((.........))))...)))))))))).(((((..(((.(......).))))))))..))))). ( -32.30) >DroGri_CAF1 82863 110 + 1 ACGG-ACG--GUG-CACGGA-CGGA-CGGUAUAGAAAGCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGGCCCUCAAAAGCGUUAUAC-UGAUGGCUAA .((.-.((--...-..))..-))..-((((((((...((.((((((((..((((.........))))...))))))))((((.....))))......)).))))))-))........ ( -31.70) >DroWil_CAF1 128269 105 + 1 A----AAGAUAUG-AACGG--CGCAAA-----AGAAAGCCAAAAGUAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGGCCGUCAAAAGCGUUCAGCUCGAAGGCUGA .----......((-(.(((--(((...-----.(....((((((((((..((((.........))))...)))))))))).)..)).))))))).......((((((....)))))) ( -28.30) >DroPer_CAF1 85052 106 + 1 A----AAG--ACG-AACGG--CCAAAG--AAGAAAAAGCCGAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGGGCUUAAAGGCCGA .----...--...-..(((--((....--.........((((((((((..((((.........))))...))))))))))...(((....)))..((((.....))))...))))). ( -30.70) >consensus A____AAG__AUG_AACGG__CCAAAG_____AAAAAGCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGAGCUUGAAGGCCGA ................(((..................(((((((((((..((((.........))))...)))))))))).)......((.(((..(((.....)))))).))))). (-16.83 = -17.25 + 0.42)

| Location | 15,122,041 – 15,122,144 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15122041 103 - 22407834 UCGGCCUUCAAGCUCAACGCUUUUGACGGUCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUCUGCUUUCGGCUUUUC-----CUUUGG--CCGUU-CAU--CUU----U ..((((.((((((.....)))..))).))))((..((....((((((.........))))))....))...))...(((((....-----....))--)))..-...--...----. ( -22.70) >DroVir_CAF1 93393 104 - 1 UUAGCCUUCA-GUUCAACGCUUUUGAGGGCCGACUGGCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGCCAU-----UCUGUCCGCCCUCUUUC-------CCCGU ..........-.....(((.....((((((.((((((((((((((((.((..(((.....)))..))..))))))))))))).-----...))).))))))....-------..))) ( -37.70) >DroPse_CAF1 83954 106 - 1 UCGGCCUUCAAGCCCAACGCUUUUGACGGUCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUCGGCUUUUUCUU--CUUUGG--CCGUU-CGU--CUU----U .(((((...(((((....(((((((..(((.....))))))))))...................((((...))))..)))))......--....))--)))..-...--...----. ( -23.82) >DroGri_CAF1 82863 110 - 1 UUAGCCAUCA-GUAUAACGCUUUUGAGGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGCUUUCUAUACCG-UCCG-UCCGUG-CAC--CGU-CCGU ..(((((..(-(((....((((....((((.....))))..((((((.........))))))..))))..))))..)))))..........-..((-..((..-...--)).-.)). ( -24.70) >DroWil_CAF1 128269 105 - 1 UCAGCCUUCGAGCUGAACGCUUUUGACGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUACUUUUGGCUUUCU-----UUUGCG--CCGUU-CAUAUCUU----U (((((......)))))........((((((.((........((((((.........))))))..(((((........)))))...-----...)))--)))))-........----. ( -22.30) >DroPer_CAF1 85052 106 - 1 UCGGCCUUUAAGCCCAACGCUUUUGACGGUCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUCGGCUUUUUCUU--CUUUGG--CCGUU-CGU--CUU----U .(((((...(((((....(((((((..(((.....))))))))))...................((((...))))..)))))......--....))--)))..-...--...----. ( -24.52) >consensus UCAGCCUUCAAGCUCAACGCUUUUGACGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUCGGCUUUUU_____CUUUCG__CCGUU_CAU__CUU____U ..((((...((((.....(((((((..(((.....))))))))))..........((((....))))....))))..)))).................................... (-15.56 = -15.67 + 0.11)


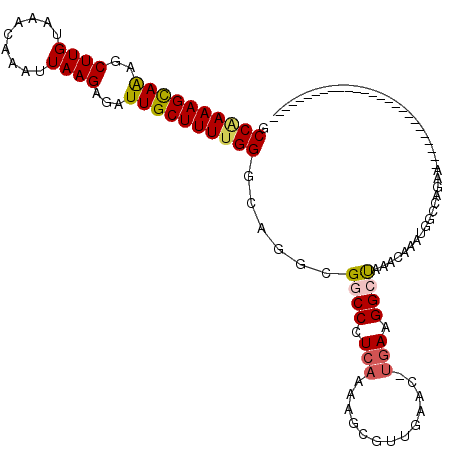
| Location | 15,122,064 – 15,122,161 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
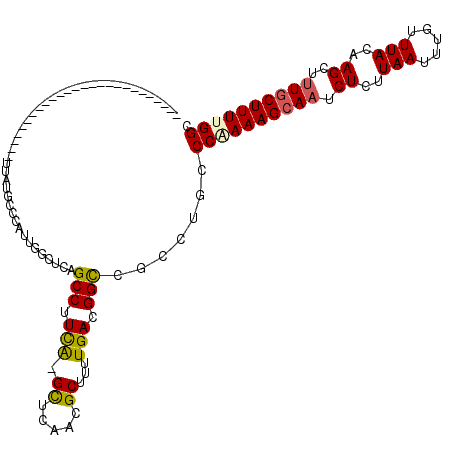
>2L_DroMel_CAF1 15122064 97 + 22407834 GCCGAAAGCAGAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGAGCUUGAAGGCCGAACAAAUGGCCAGAACAG----------------------- .((((((((((..((((.........))))...)))))))))).(((((...(((.....)))....)))))..(((((......))))).......----------------------- ( -29.40) >DroVir_CAF1 93418 95 + 1 GCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGCCAGUCGGCCCUCAAAAGCGUUGAAC-UGAAGGCUAAGCCAAUGGGCGUA---------------GC---------G (((((((((((..((((.........))))...)))))))))))((((((((.((....)).)))).))-))...((((.(((....))).))---------------))---------. ( -34.50) >DroGri_CAF1 82894 119 + 1 GCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGGCCCUCAAAAGCGUUAUAC-UGAUGGCUAAGCCAAUGGGCACAUCAGAUCUGAGCGGAGCGAGCGCGGUG (((((((((((..((((.........))))...))))))))((((.....))))......(((((...(-(((((.....(((....))).)))))).(((....)))...)))))))). ( -37.00) >DroWil_CAF1 128294 94 + 1 GCCAAAAGUAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGGCCGUCAAAAGCGUUCAGCUCGAAGGCUGAACAAAUGCCCAAAA-------------------------- ...((((((((..((((.........))))...))))))))((((((((....)))......((((((((....))))))))...)))))....-------------------------- ( -30.40) >DroMoj_CAF1 93620 94 + 1 GCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUCGGCUAGGCGGCCCUCAAAAGCGUUGAAC-UGAAGGCUAAGCCAAUGGGCGUG---------------GC---------- (((((((((((..((((.........))))...)))))))((.((((((((((.(((............-))).))))..)))..))).))))---------------))---------- ( -26.10) >DroPer_CAF1 85078 97 + 1 GCCGAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGACCGUCAAAAGCGUUGGGCUUAAAGGCCGAACAAAUGGCCAGAACGG----------------------- .((((((((((..((((.........))))...))))))))))........(((((..((((.....))))...(((((......))))).).))))----------------------- ( -29.50) >consensus GCCAAAAGCAAAGCUUGUAAACAAAUUAAGAGAUUGCUUUUGGGCAGGCGGCCCUCAAAAGCGUUGAAC_UGAAGGCUAAACAAAUGGCCAGAA__________________________ .((((((((((..((((.........))))...))))))))))......((((.(((.............))).)))).......................................... (-17.03 = -17.20 + 0.17)

| Location | 15,122,064 – 15,122,161 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15122064 97 - 22407834 -----------------------CUGUUCUGGCCAUUUGUUCGGCCUUCAAGCUCAACGCUUUUGACGGUCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUCUGCUUUCGGC -----------------------.......((((........))))...((((.....(((((((..(((.....))))))))))..........((((....))))....))))..... ( -19.90) >DroVir_CAF1 93418 95 - 1 C---------GC---------------UACGCCCAUUGGCUUAGCCUUCA-GUUCAACGCUUUUGAGGGCCGACUGGCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGC .---------((---------------((.(((....))).))))...((-(((....((((....)))).)))))(((((((((((.((..(((.....)))..))..))))))))))) ( -30.50) >DroGri_CAF1 82894 119 - 1 CACCGCGCUCGCUCCGCUCAGAUCUGAUGUGCCCAUUGGCUUAGCCAUCA-GUAUAACGCUUUUGAGGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGC ......((..((.((.((((((..((.(((((....((((...))))...-))))).))..))))))))..))..))((((((((((.((..(((.....)))..))..)))))))))). ( -34.60) >DroWil_CAF1 128294 94 - 1 --------------------------UUUUGGGCAUUUGUUCAGCCUUCGAGCUGAACGCUUUUGACGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUACUUUUGGC --------------------------..(((((((...(((((((......)))))))(((......)))....)))))))((((((.........))))))....(((........))) ( -26.00) >DroMoj_CAF1 93620 94 - 1 ----------GC---------------CACGCCCAUUGGCUUAGCCUUCA-GUUCAACGCUUUUGAGGGCCGCCUAGCCGAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGC ----------((---------------((.......))))...(((((((-(..........))))))))......(((((((((((.((..(((.....)))..))..))))))))))) ( -30.00) >DroPer_CAF1 85078 97 - 1 -----------------------CCGUUCUGGCCAUUUGUUCGGCCUUUAAGCCCAACGCUUUUGACGGUCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUCGGC -----------------------(((((..((((........))))...((((.....))))..)))))......(((..(((((((.((..(((.....)))..))..))))))).))) ( -23.30) >consensus __________________________UUAUGCCCAUUGGCUCAGCCUUCA_GCUCAACGCUUUUGACGGCCGCCUGCCCAAAAGCAAUCUCUUAAUUUGUUUACAAGCUUUGCUUUUGGC ...........................................(((.(((.((.....))...))).))).......((((((((((.((..(((.....)))..))..)))))))))). (-15.43 = -15.27 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:47 2006