
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,120,598 – 15,120,693 |
| Length | 95 |
| Max. P | 0.582547 |

| Location | 15,120,598 – 15,120,693 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15120598 95 - 22407834 -CCCGA---------UGCCUUUAU------AUU--UCUGUUUUGUGUUUACAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC-------UCCUGGGCAGGACAG -.((..---------((((((...------(((--((..(((((((....)))))))..)))))))))))...(((.(..((((((........))))-------))..).))))).... ( -33.40) >DroVir_CAF1 91336 94 - 1 -CUUUA---------UGCCUUUAU------AU---UCUGUUUUGUGUUUACAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC-------UUCUGGGCAGGGAUA -.....---------((((((...------.(---((..(((((((....)))))))..)))..))))))..((((.(..((((((........))))-------))..).))))..... ( -30.00) >DroPse_CAF1 82147 102 - 1 GCUCUG---------UGCCUUUAUGCUCGUAUU--UCUGUUUUGUGUUUGCAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC-------UCCUGGGCAGGGCAG (((((.---------.((.....((((..((((--((..(((((((....)))))))..)))))).))))....))((((((((((........))))-------))..))))))))).. ( -37.00) >DroGri_CAF1 81416 94 - 1 -CUUUA---------UGCCUUUAU------AU---UCUGUUUUGUGUUUACAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC-------UUCUGGGCAGGGAUA -.....---------((((((...------.(---((..(((((((....)))))))..)))..))))))..((((.(..((((((........))))-------))..).))))..... ( -30.00) >DroWil_CAF1 126499 113 - 1 -CUCUGUGCUUUUGCUGUCUCUAU------UUUAUUCUGUUUUGUGUUUACAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGACUCGUCGUCCUGGGAAGGGAGG -(((.((((.....(((((((.((------(((((....(((((((....)))))))))))))))))))))...)))).(((((.((((....)))))))))....(((....)))))). ( -36.70) >DroPer_CAF1 83267 102 - 1 GCUCUG---------UGCCUUUAUGCUCGUAUU--UCUGUUUUGUGUUUGCAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC-------UCCUGGGCAGGGCAG (((((.---------.((.....((((..((((--((..(((((((....)))))))..)))))).))))....))((((((((((........))))-------))..))))))))).. ( -37.00) >consensus _CUCUA_________UGCCUUUAU______AUU__UCUGUUUUGUGUUUACAUAAAAGUGAAGUGAGGCAGAUUGCGCUCGAGUGCGUUAAUCAGCGC_______UCCUGGGCAGGGAAG ...............(((((...............(((((((..(.(((((......))))))..)))))))..(((((.((.........)))))))...........)))))...... (-22.08 = -22.05 + -0.03)

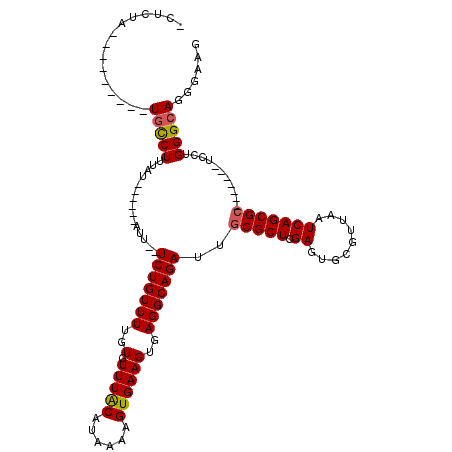
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:44 2006