| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,091,404 – 15,091,508 |
| Length | 104 |
| Max. P | 0.508942 |

| Location | 15,091,404 – 15,091,508 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.03 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
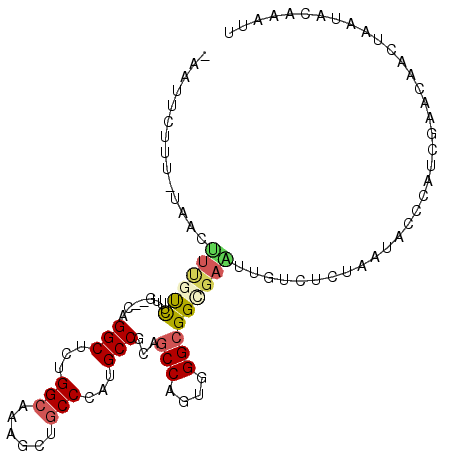
>2L_DroMel_CAF1 15091404 104 + 22407834 -AAUUCUUUCUAACUUUGUCUUUG--UAGGCUCUGGCCAAGUUGCCCAUGCCACAGCCAGUGGGCGGCGAGUUGUCUCUUAUACCCAUCGAACAACUAAUACAAAUU -...................((((--(((((....)))..(((((((((((....))..))))))))).((((((.((...........))))))))..)))))).. ( -27.70) >DroVir_CAF1 52115 103 + 1 GUAUACUCU-UUUUCAA-UUUUUG--CAGGCUCUGGCUAAGCUGCCAAUGCCUCAGCCCGUGGGCGGUGAACUGUCCUUAAUACCCAUCGAGCAACUAAUACAAAUU ((((.....-.......-....((--.((((..((((......))))..))))))((.((.((((((....))))))...........)).)).....))))..... ( -28.30) >DroEre_CAF1 36775 104 + 1 -AAUUCCUGGUAACUUCGUCUUUG--CAGGCUCUGGCAAAGCUGCCCAUGCCGCAGCCCGUGGGCGGCGAAUUGUCGCUUAUACCCAUCGAACAACUUAUACAAAUU -.......((((.....((((...--.))))...(((((.((((((((((........))))))))))...))))).....))))...................... ( -29.00) >DroWil_CAF1 75697 104 + 1 -AAUUAAUUUUAAUUUUGUUUUUU--UAGGCACUGGCCAAGUUACCCAUGCCUCAGCCAGUGGGAGGUGAAUUAUCUUUAAUACCCAUCGAGCAAUUAAUACAAAUU -.(((((((...............--.(((((.(((.........))))))))..(((.(((((...((((.....))))...))))).).)))))))))....... ( -19.80) >DroYak_CAF1 43922 104 + 1 -AAUUCUUAAUAACUUCGUCUAUG--CAGGCUCUGGCAAAGCUGCCCAUGCCGCAGCCAGUUGGCGGCGAAUUGUCUCUUAUACCCAUCGAACAACUCAUACAAAUU -........((((.((((((..((--(.(((...((((....))))...))))))(((....)))))))))))))................................ ( -25.70) >DroMoj_CAF1 47675 105 + 1 GAAUUCUAA-UUCUCUU-CUCUUACCCAGGCUCUGGCUAAGCUGCCAAUGCCGCAGCCCGUGGGCGGUGAGCUGUCUCUAAUACCCAUCGAACAUCUAAUACAAAUC ((((....)-)))....-..((((((..(((..((((......))))..)))...(((....))))))))).................................... ( -26.10) >consensus _AAUUCUUU_UAACUUUGUCUUUG__CAGGCUCUGGCAAAGCUGCCCAUGCCGCAGCCAGUGGGCGGCGAAUUGUCUCUAAUACCCAUCGAACAACUAAUACAAAUU ..............((((((........(((...(((......)))...)))...(((....))))))))).................................... (-15.03 = -15.28 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:33 2006