| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,067,832 – 15,067,951 |
| Length | 119 |
| Max. P | 0.811185 |

| Location | 15,067,832 – 15,067,951 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
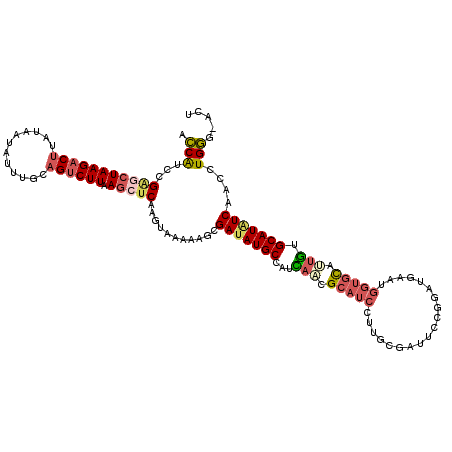
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15067832 119 + 22407834 AGU-CCCAGGUUGAUAUGCACAAUGCACCAUUCAUCCGAAAUCGCAAGGAUGCGUUGAUGGCAUAUCGCUUUUUACUCGAGCUUAAGACUGCAAAUAUUAUAAGUCUUAGCUCGGAUGGU .((-((.((((.(((((((.(((((((...(((....))).((....)).)))))))...))))))))))).......(((((.((((((............)))))))))))))))... ( -39.00) >DroGri_CAF1 11989 119 + 1 AGCCAACA-GUAGACAUGCGUUCCAAUGACUUUAUCAAGACUCGCAUGGAGGCGGCAAUUGCAUUUCGAGUGCUGCUCGAUUUGAAGGCGGCCAACAUUGUUAGUCUUAGCGCUGAUGCU .......(-(((..((.(((((.....((.....))((((((.(((((..(((.((.....((..(((((.....)))))..))...)).)))..)).))).))))))))))))).)))) ( -37.30) >DroSec_CAF1 12569 119 + 1 AGU-CCCAGGUUGAUAUGCACAAUGCACCAUUCAUUCGCAAUCGCAAAGAUGCGUUGAUGGCAUAUCGCUUUUUACUUGAGCUUAAGACUGCAAAUAUAAUAAGUCUUAGCUCGGAUGGU .((-((.((((.(((((((.(((((((....((.((.((....)).)))))))))))...))))))))))).......(((((.((((((............)))))))))))))))... ( -36.70) >DroSim_CAF1 9686 119 + 1 AGU-CCCAGGUUGAUAUGCACAAUGCACCAUUCAUUCGCAAUCGCAAGGAUGCGUUGAUGGCAUAUCGCUUUUUACUUGAGCUUAAGACUGCAAAUAUUAUAAGUCUUAGCUCGGAUGGU .((-((.((((.(((((((.(((((((((..................)).)))))))...))))))))))).......(((((.((((((............)))))))))))))))... ( -37.57) >DroEre_CAF1 12486 119 + 1 AGU-CCCAGGUUGAUAUGCACAAUGCACCCUUCAUCCGGAACCGCCAAGAUGCGGUGAUGGCAUAUCGCUUUAUACUUGAGCUUAAGACUGCAAAUGUUAUAAGUCUUAGCCCGGACGGU .((-((..(((.(((((((.((...((((...((((.((.....))..)))).)))).)))))))))((((.......)))).(((((((............)))))))))).))))... ( -37.30) >DroYak_CAF1 12120 119 + 1 AGU-GCCAGGUGGACAUGCACAAUGCACCUUUUAUCCGUAAUCGCAAAGAUGCGAUAAUGGCAUAUCGCUUUUUACUUGAGCUCAAGACUGCAAAUAUUAUAAGUCUUAGUCCGGACGGU ...-(((...(((((.(((.....))).....(((((((.((((((....)))))).)))).)))..((((.......))))..((((((............)))))).)))))...))) ( -30.50) >consensus AGU_CCCAGGUUGAUAUGCACAAUGCACCAUUCAUCCGCAAUCGCAAAGAUGCGUUGAUGGCAUAUCGCUUUUUACUUGAGCUUAAGACUGCAAAUAUUAUAAGUCUUAGCUCGGAUGGU .....((.(((.(((((((.(((((((.......................)))))))...)))))))((((.......)))).(((((((............)))))))))).))..... (-19.68 = -20.52 + 0.84)

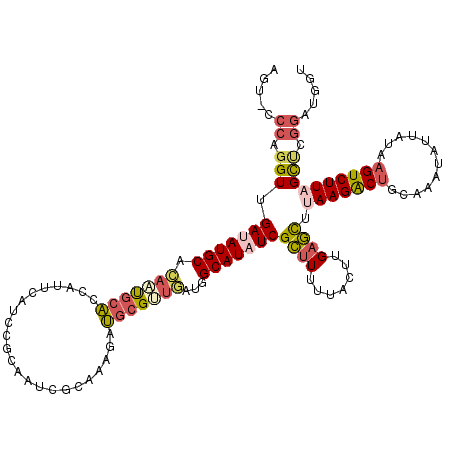
| Location | 15,067,832 – 15,067,951 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15067832 119 - 22407834 ACCAUCCGAGCUAAGACUUAUAAUAUUUGCAGUCUUAAGCUCGAGUAAAAAGCGAUAUGCCAUCAACGCAUCCUUGCGAUUUCGGAUGAAUGGUGCAUUGUGCAUAUCAACCUGGG-ACU .(((..((((((((((((((.......)).)))))).))))))..........((((((((((((...(((((..........)))))..)))))......)))))))....))).-... ( -35.50) >DroGri_CAF1 11989 119 - 1 AGCAUCAGCGCUAAGACUAACAAUGUUGGCCGCCUUCAAAUCGAGCAGCACUCGAAAUGCAAUUGCCGCCUCCAUGCGAGUCUUGAUAAAGUCAUUGGAACGCAUGUCUAC-UGUUGGCU (((......))).((.((((((.....(((.((...((..(((((.....)))))..)).....)).)))..((((((..((.((((...))))..))..)))))).....-)))))))) ( -29.40) >DroSec_CAF1 12569 119 - 1 ACCAUCCGAGCUAAGACUUAUUAUAUUUGCAGUCUUAAGCUCAAGUAAAAAGCGAUAUGCCAUCAACGCAUCUUUGCGAUUGCGAAUGAAUGGUGCAUUGUGCAUAUCAACCUGGG-ACU ....((((((((((((((((.......)).)))))).)))))...........(((((((...(((.((((((((((....))))).....))))).))).)))))))......))-).. ( -35.70) >DroSim_CAF1 9686 119 - 1 ACCAUCCGAGCUAAGACUUAUAAUAUUUGCAGUCUUAAGCUCAAGUAAAAAGCGAUAUGCCAUCAACGCAUCCUUGCGAUUGCGAAUGAAUGGUGCAUUGUGCAUAUCAACCUGGG-ACU ....((((((((((((((((.......)).)))))).)))))...........(((((((...(((.(((((.((((....))))......))))).))).)))))))......))-).. ( -33.90) >DroEre_CAF1 12486 119 - 1 ACCGUCCGGGCUAAGACUUAUAACAUUUGCAGUCUUAAGCUCAAGUAUAAAGCGAUAUGCCAUCACCGCAUCUUGGCGGUUCCGGAUGAAGGGUGCAUUGUGCAUAUCAACCUGGG-ACU ...(((((((((((((((((.......)).)))))).)))))...........(((((((((.((((.(((((.((.....)))))))...))))...)).)))))))......))-)). ( -40.30) >DroYak_CAF1 12120 119 - 1 ACCGUCCGGACUAAGACUUAUAAUAUUUGCAGUCUUGAGCUCAAGUAAAAAGCGAUAUGCCAUUAUCGCAUCUUUGCGAUUACGGAUAAAAGGUGCAUUGUGCAUGUCCACCUGGC-ACU ...(((((((((((((((((.......)).)))))).)).))..(((((..((((((......))))))...))))).....)))))....(((((...(((......)))...))-))) ( -31.50) >consensus ACCAUCCGAGCUAAGACUUAUAAUAUUUGCAGUCUUAAGCUCAAGUAAAAAGCGAUAUGCCAUCAACGCAUCCUUGCGAUUCCGGAUGAAUGGUGCAUUGUGCAUAUCAACCUGGG_ACU .(((...(((((((((((............)))))).)))))...........(((((((...(((.(((((...................))))).))).)))))))....)))..... (-21.95 = -22.84 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:27 2006