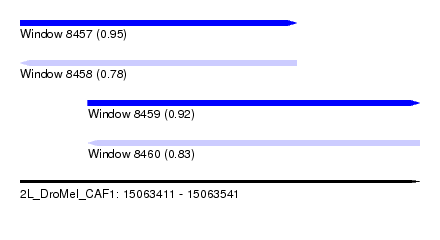
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,063,411 – 15,063,541 |
| Length | 130 |
| Max. P | 0.952140 |

| Location | 15,063,411 – 15,063,501 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -20.78 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
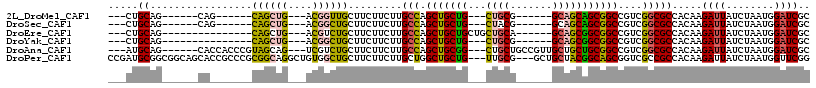
>2L_DroMel_CAF1 15063411 90 + 22407834 ---CUGCAG------CAG------CAGCUG---ACGGUUGCUUCUUCUUGCCAGCUGCUG---CUGCG------GCAGCAGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC ---.(((..------..)------))((((---((((((((........))..(((((((---(....------)))))))))))))))))).(((..((.....))..)))..... ( -39.90) >DroSec_CAF1 7750 90 + 1 ---CUGCAG------CAG------CAGCUG---ACGGCUGCUUCUUCUUGCCAGCUGCUG---CUACG------GCAGCAGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC ---.(((..------..)------))((((---((((((((........))..(((((((---(....------)))))))))))))))))).(((..((.....))..)))..... ( -44.90) >DroEre_CAF1 8249 90 + 1 ---CUGCAG---------------CAGCUG---ACGUCUGCUUCUUCUUGCCAGCUGCUGCUGCUGCA------GCAGCGGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC ---.....(---------------(.((((---(((...((........))..(((((((((((....------)))))))))))))))))).(((..((.....))..)))...)) ( -38.40) >DroYak_CAF1 7915 87 + 1 ---CUGCAG---------------CAGCUG---ACGGCUGCUUCUUCUUGCCAGCUGCUG---CUGCG------GCAGCGGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC ---.....(---------------(.((((---((((((((........))..(((((((---(....------)))))))))))))))))).(((..((.....))..)))...)) ( -40.90) >DroAna_CAF1 8696 102 + 1 ---AUGCAG------CACCACCCGUAGCAG---UCGUCUGCUUCUUCUUGCCAGCUGCGG---CUGCUGCCGUUGCUGCUGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC ---..((((------((.((.(.(((((((---((((..(((..........))).))))---))))))).).)).))))))((((....).)))....((((........)))).. ( -39.30) >DroPer_CAF1 7500 111 + 1 CCGAUGCGGCGGCAGCACCGCCCGCGGCAGGCUGUGGCUGCUUCUUCUUGCUGGCUGCUG---UUGCG---GCUGCUACGGCAGCGGUCGCCGCCACAAGAUUAUCUAAUGGUUCGG ((((.(((((.(((((.(((....)))...))))).)))))....(((((.((((.((..---..((.---(((((....))))).)).)).)))))))))............)))) ( -54.90) >consensus ___CUGCAG______CA_______CAGCUG___ACGGCUGCUUCUUCUUGCCAGCUGCUG___CUGCG______GCAGCGGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGC .....((.................((((((....)))))).........(((.(((((((...((((.......)))))))))))....))))).....((((........)))).. (-20.78 = -20.73 + -0.05)
| Location | 15,063,411 – 15,063,501 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -41.08 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15063411 90 - 22407834 GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCUGCUGC------CGCAG---CAGCAGCUGGCAAGAAGAAGCAACCGU---CAGCUG------CUG------CUGCAG--- (((..((((.((.....)).))))))).(.((((....)))).------)((((---(((((((((((..............))---))))))------)))------))))..--- ( -42.24) >DroSec_CAF1 7750 90 - 1 GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCUGCUGC------CGUAG---CAGCAGCUGGCAAGAAGAAGCAGCCGU---CAGCUG------CUG------CUGCAG--- (((..((((.((.....)).))))))).((((((.((((((((------....)---)))))))..((........)).)))))---)..(((------(..------..))))--- ( -40.30) >DroEre_CAF1 8249 90 - 1 GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCCGCUGC------UGCAGCAGCAGCAGCUGGCAAGAAGAAGCAGACGU---CAGCUG---------------CUGCAG--- ((..............((((((((((((...)).))))(((((------(((....))))))))..))))))...(((((.(..---..))))---------------))))..--- ( -39.20) >DroYak_CAF1 7915 87 - 1 GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCCGCUGC------CGCAG---CAGCAGCUGGCAAGAAGAAGCAGCCGU---CAGCUG---------------CUGCAG--- ((((((.....)))......((((((((...)).))))))...------))).(---(((((((((((..............))---))))))---------------))))..--- ( -38.54) >DroAna_CAF1 8696 102 - 1 GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCAGCAGCAACGGCAGCAG---CCGCAGCUGGCAAGAAGAAGCAGACGA---CUGCUACGGGUGGUG------CUGCAU--- (((..((((..(....((((((.(((((...)).)))..((((..((((....)---)))..))))))))))...(((((....---))))).)..))))..------.)))..--- ( -38.00) >DroPer_CAF1 7500 111 - 1 CCGAACCAUUAGAUAAUCUUGUGGCGGCGACCGCUGCCGUAGCAGC---CGCAA---CAGCAGCCAGCAAGAAGAAGCAGCCACAGCCUGCCGCGGGCGGUGCUGCCGCCGCAUCGG ((((..(((.((.....)).)))((((((...(((((.((.((...---.)).)---).)))))............(((((.((.(((((...))))).))))))))))))).)))) ( -48.20) >consensus GCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCCGCUGC______CGCAG___CAGCAGCUGGCAAGAAGAAGCAGCCGU___CAGCUG_______UG______CUGCAG___ (((..((((.((.....)).))))))).....(((((.((((...............)))).)).)))........((((............................))))..... (-18.92 = -18.90 + -0.02)
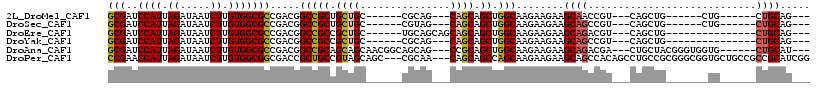
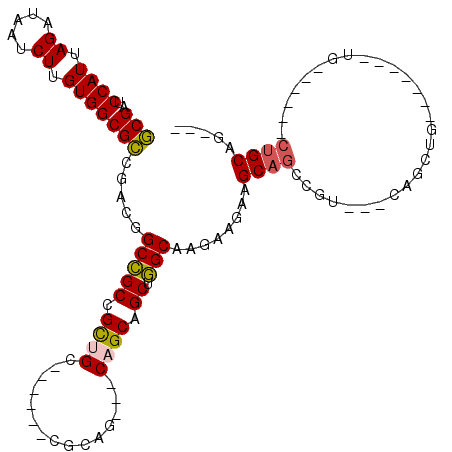
| Location | 15,063,433 – 15,063,541 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -20.97 |
| Energy contribution | -22.05 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15063433 108 + 22407834 CUUCUUCUUGCCAGCUGCUG---CUGCG------GCAGCAGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGCUGACAUCGUCGUCCAUGAACAAAAGUUCAUAACGAUUUUU .........(((.(((((((---(....------))))))))))).((((((((((..((.....))..)))..)))))))...(((((..((((((....)))))).))))).... ( -45.20) >DroPse_CAF1 7247 111 + 1 CUUCUUCUUGCUGGCUGCUG---UUGCG---GCUGCUACGGCAGCGGUCGCCGCCACAAGAUUAUCUAAUGGUUCGGAAACAUCAUCGUCCAUGAACAAGAGUUCAUAACGAUUUUU .....(((((.((((.((..---..((.---(((((....))))).)).)).)))))))))..............(....)...(((((..((((((....)))))).))))).... ( -38.80) >DroEre_CAF1 8268 111 + 1 CUUCUUCUUGCCAGCUGCUGCUGCUGCA------GCAGCGGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGCUGACAUCGUCGUCCAUGAACAAAAGUUCAUAACGAUUUUU .........(((.((((((((((...))------))))))))))).((((((((((..((.....))..)))..)))))))...(((((..((((((....)))))).))))).... ( -46.80) >DroMoj_CAF1 7023 91 + 1 -------UUGAUGGCUGCUG---CUUCU----------------UCUUGGCCGCGACAAGAUUAUCUAAUGGAUCGCCAACAUCGUCGUCCAUGAACAAGAGUUCAUAACGAUUUUU -------.((.(((((((.(---((...----------------....))).)))....((((........)))))))).))..(((((..((((((....)))))).))))).... ( -23.60) >DroAna_CAF1 8724 114 + 1 CUUCUUCUUGCCAGCUGCGG---CUGCUGCCGUUGCUGCUGCGGCCGUCGGCGCCACAAGAUUAUCUAAUGGAUCGCUGACAUCGUCGUCCAUGAACAAGAGCUCAUAACGAUUUUU ....((((((.((((.((((---(....))))).))))..(((((.((((((((((..((.....))..)))..)))))))...))))).......))))))............... ( -38.30) >DroPer_CAF1 7540 111 + 1 CUUCUUCUUGCUGGCUGCUG---UUGCG---GCUGCUACGGCAGCGGUCGCCGCCACAAGAUUAUCUAAUGGUUCGGAAACAUCAUCGUCCAUGAACAAGAGUUCAUAACGAUUUUU .....(((((.((((.((..---..((.---(((((....))))).)).)).)))))))))..............(....)...(((((..((((((....)))))).))))).... ( -38.80) >consensus CUUCUUCUUGCCAGCUGCUG___CUGCG______GCUGCGGCGGCCGUCGCCGCCACAAGAUUAUCUAAUGGAUCGCUAACAUCGUCGUCCAUGAACAAGAGUUCAUAACGAUUUUU .............(((((((.....((.......))..))))))).((.(((((((..((.....))..)))..)))).))...(((((..((((((....)))))).))))).... (-20.97 = -22.05 + 1.08)

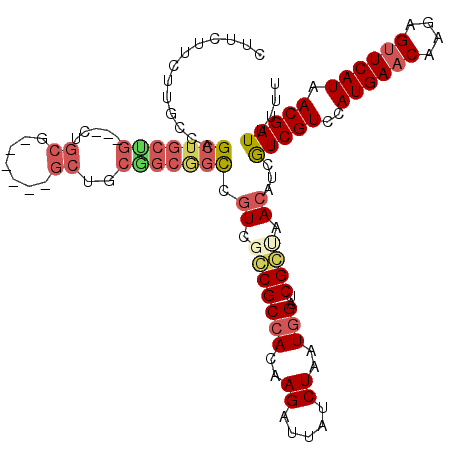
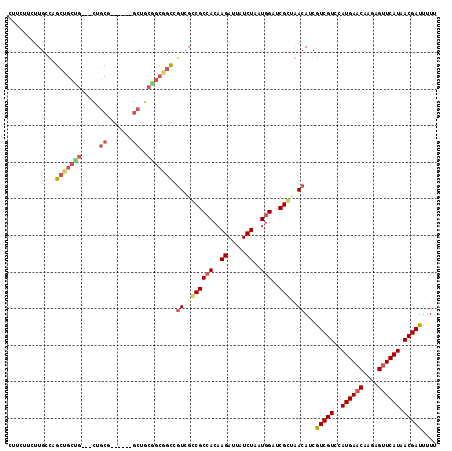
| Location | 15,063,433 – 15,063,541 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15063433 108 - 22407834 AAAAAUCGUUAUGAACUUUUGUUCAUGGACGACGAUGUCAGCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCUGCUGC------CGCAG---CAGCAGCUGGCAAGAAGAAG .....(((((((((((....)))))).)))))...((((.(((..((((.((.....)).))))))).))))(((((((((((------....)---))))))).)))......... ( -44.80) >DroPse_CAF1 7247 111 - 1 AAAAAUCGUUAUGAACUCUUGUUCAUGGACGAUGAUGUUUCCGAACCAUUAGAUAAUCUUGUGGCGGCGACCGCUGCCGUAGCAGC---CGCAA---CAGCAGCCAGCAAGAAGAAG ....((((((((((((....)))))).))))))...(((.(((..((((.((.....)).))))))).))).(((((.((.((...---.)).)---).)))))............. ( -34.10) >DroEre_CAF1 8268 111 - 1 AAAAAUCGUUAUGAACUUUUGUUCAUGGACGACGAUGUCAGCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCCGCUGC------UGCAGCAGCAGCAGCUGGCAAGAAGAAG .....(((((((((((....)))))).)))))...((((.(((..((((.((.....)).))))))).))))...((((((((------(((....)))))))).)))......... ( -45.60) >DroMoj_CAF1 7023 91 - 1 AAAAAUCGUUAUGAACUCUUGUUCAUGGACGACGAUGUUGGCGAUCCAUUAGAUAAUCUUGUCGCGGCCAAGA----------------AGAAG---CAGCAGCCAUCAA------- .....(((((((((((....)))))).)))))...((.((((.........((((....))))((.((.....----------------....)---).)).)))).)).------- ( -23.10) >DroAna_CAF1 8724 114 - 1 AAAAAUCGUUAUGAGCUCUUGUUCAUGGACGACGAUGUCAGCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCAGCAGCAACGGCAGCAG---CCGCAGCUGGCAAGAAGAAG .....(((((((((((....)))))).)))))...(((((((...((((.((.....)).))))....(.((((.((.((.......)).)).)---)))).)))))))........ ( -38.20) >DroPer_CAF1 7540 111 - 1 AAAAAUCGUUAUGAACUCUUGUUCAUGGACGAUGAUGUUUCCGAACCAUUAGAUAAUCUUGUGGCGGCGACCGCUGCCGUAGCAGC---CGCAA---CAGCAGCCAGCAAGAAGAAG ....((((((((((((....)))))).))))))...(((.(((..((((.((.....)).))))))).))).(((((.((.((...---.)).)---).)))))............. ( -34.10) >consensus AAAAAUCGUUAUGAACUCUUGUUCAUGGACGACGAUGUCAGCGAUCCAUUAGAUAAUCUUGUGGCGCCGACGGCCGCCGCAGC______CGCAG___CAGCAGCCAGCAAGAAGAAG .....(((((((((((....)))))).)))))...(((((((..................(((((.......))))).............((.......)).)))))))........ (-22.70 = -23.57 + 0.86)

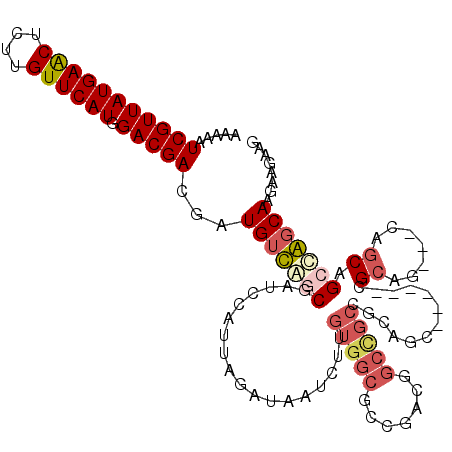
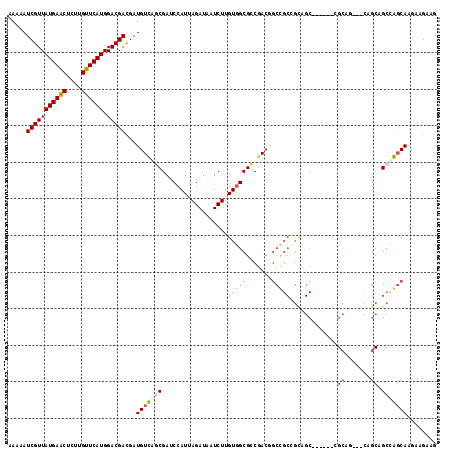
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:25 2006