
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,038,983 – 15,039,089 |
| Length | 106 |
| Max. P | 0.989366 |

| Location | 15,038,983 – 15,039,089 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -7.95 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
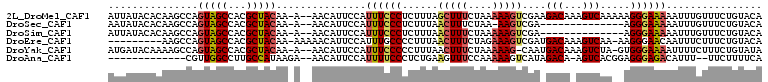
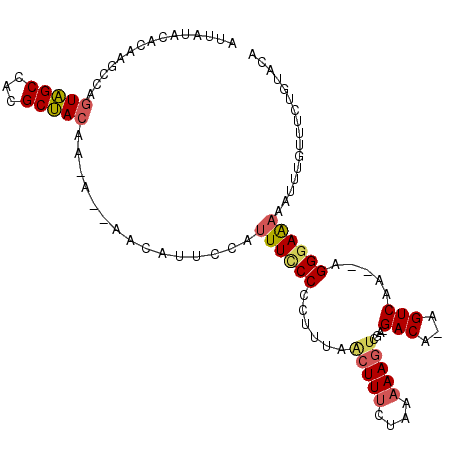
>2L_DroMel_CAF1 15038983 106 + 22407834 AUUAUACACAAGCCAGUAGCCACGCUACAA-A--AACAUUCCAUUUCCCUCUUUAGCUUUCUAAAAAGUCGAAGACAAAGUCAAAAAGGGAAAAAUUUGUUUCUGUACA ....((((.((((..(((((...)))))..-.--.........((((((((((..(((((....)))))..))))............)))))).....)))).)))).. ( -22.30) >DroSec_CAF1 7513 91 + 1 AAUAUACACAAGCCAGUAGCCACGCUACAA-A--AACAUUCCAUUUCCCUCUUUAACUUUCUAA-AAGUCGA--------------AGGGAAAAAUUUGUUUCUGUACA ....((((.((((..(((((...)))))..-.--.........(((((((..(..((((.....-))))..)--------------))))))).....)))).)))).. ( -19.70) >DroSim_CAF1 7567 92 + 1 AUUAUACACAAGCCAGUAGCCACGCUACAA-A--AACAUUCCAUUUCCCUCUUUAACUUUCUAAAAAGUCGA--------------AGGGAAAAAUUUGUUUCUGUACA ....((((.((((..(((((...)))))..-.--.........(((((((..(..(((((....)))))..)--------------))))))).....)))).)))).. ( -20.50) >DroEre_CAF1 7288 98 + 1 ---------AAGCCAGUAGCCACGCUACAA-AAAAACAUUCCAUUUGCCCCUUUAACUUUCUAGAAAGUCGAUGACAAAGUCAA-AAGGGAACAAUUUCUUUCUGUACA ---------......(((((...)))))..-.................((((((.(((((....)))))...((((...)))))-)))))................... ( -16.20) >DroYak_CAF1 7400 104 + 1 AUGAUACAAAAGCCAGUAGCCACGCUACAA-A--AACAUUCCAUUUCCCCCUUUAACUUUCUAAAAAG-CAAUGACAAAGUCUA-GUGGGAAAAUUUUCUUUCUGUAUA ...(((((((((...(((((...)))))..-.--.........((((((.((...(((((........-.......)))))..)-).)))))).....)))).))))). ( -18.66) >DroAna_CAF1 7447 91 + 1 -------------CGUUGGCCUUGCCAUAAGA--AACAUUCCAUUUUCCCUCUGAAGUUUCCAAAAAGUCAUAGACA-AGUCACGGAGGGAGACAUUU--UUCUUUUCA -------------...((((...))))...((--((........((((((((((..(....).....(((...))).-.....)))))))))).....--....)))). ( -18.53) >consensus AUUAUACACAAGCCAGUAGCCACGCUACAA_A__AACAUUCCAUUUCCCCCUUUAACUUUCUAAAAAGUCGA_GACA_AGUCAA__AGGGAAAAAUUUGUUUCUGUACA ...............(((((...)))))...............((((((......(((((....)))))....(((...))).....))))))................ ( -7.95 = -8.57 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:15 2006