
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
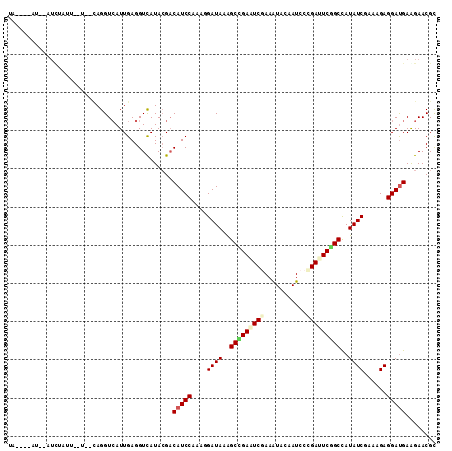
| Location | 15,026,657 – 15,026,802 |
| Length | 145 |
| Max. P | 0.975813 |

| Location | 15,026,657 – 15,026,762 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15026657 105 + 22407834 UG----AU--CUCUAUUC-GCACAGGUCAUUGAGGUUAUACGUCAUCCCAAGGAUAAGGCCGAAUCGAAAUACAAUCCCGAUUCAGCCAUAUCGAAAGAGGACGAAGAACGC ((----((--(((...((-.....)).....)))))))..((((((((...))))..(((.((((((...........)))))).)))...((....)).))))........ ( -27.10) >DroGri_CAF1 18883 108 + 1 UA----AUAUAUCAAUUGAUCUUAGGUCAUUGAGGUCAUACGACAUCCAAAGGAUAAGGCCGAAUCAAAGUACAAUCCGGACUCGGCCAUAUCGAAGGAGGAUGAGGAACGC ..----.....((...(((((((((....)))))))))...))(((((.........((((((.((...(......)..)).))))))...((....)))))))........ ( -30.00) >DroWil_CAF1 34216 106 + 1 UAAAAACU--AUUUUUU--U--AAGGUCAUUGAGGUCAUAAGACAUCCUAAAGAUAAAGCUGAAUCGAAAUAUAAUCCCGAUUCGGCUAUAUCGAAAGAGGAUGAAGAACGU ........--....(((--(--(.(..(.....)..).)))))((((((...((((.((((((((((...........)))))))))).)))).....))))))........ ( -30.20) >DroMoj_CAF1 22854 105 + 1 GU----AUAUAUUAAU---UUUCAGGUCAUUGAGGUCAUACGACAUCCAAAAGAUAAAGCCGAAUCCAAGUAUAAUCCAGACUCGGCCAUAUCCAAAGAGGAUGAGGAACGC ..----..........---((((.(..(.....)..)......(((((....((((..(((((.((.............)).)))))..))))......))))).))))... ( -19.42) >DroAna_CAF1 19813 99 + 1 AC----CA--AUCUC-------UAGGUAAUUGAGGUCAUACGACAUCCGAAGGAUAAAGCUGAAUCGAAGUAUAAUCCUGAUUCAGCCAUAUCGAAAGAGGAUGAGGAACGC .(----(.--(((((-------.........))))).......(((((..........(((((((((...........)))))))))....((....))))))).))..... ( -26.20) >DroPer_CAF1 31407 99 + 1 UG----CC--U----UUC-G--CAGGUCAUUGAGGUCAUACGACAUCCCAAGGAUAAAGCCGAGUCGAAAUACAAUCCCGAUUCGGCCAUAUCGAAGGAGGAUGAAGAACGC .(----((--(----(..-.--.........)))))....(..((((((...((((..(((((((((...........)))))))))..))))....).)))))..)..... ( -27.80) >consensus UA____AU__AUCUAUU__U__CAGGUCAUUGAGGUCAUACGACAUCCAAAGGAUAAAGCCGAAUCGAAAUACAAUCCCGAUUCGGCCAUAUCGAAAGAGGAUGAAGAACGC ...........................................(((((....((((..(((((((((...........)))))))))..))))......)))))........ (-18.39 = -18.95 + 0.56)


| Location | 15,026,657 – 15,026,762 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.55 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
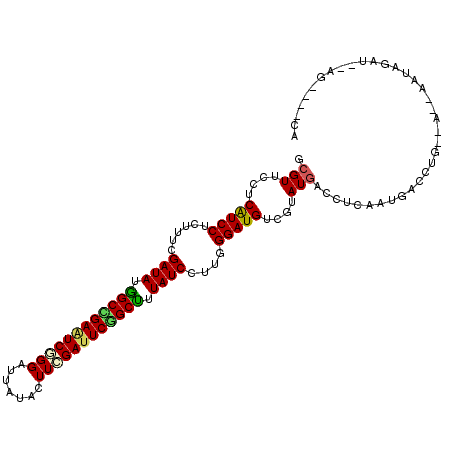
>2L_DroMel_CAF1 15026657 105 - 22407834 GCGUUCUUCGUCCUCUUUCGAUAUGGCUGAAUCGGGAUUGUAUUUCGAUUCGGCCUUAUCCUUGGGAUGACGUAUAACCUCAAUGACCUGUGC-GAAUAGAG--AU----CA ..(.((((((((((.....((((.((((((((((..((...))..)))))))))).))))...)))))))((((((............)))))-).....))--).----). ( -35.30) >DroGri_CAF1 18883 108 - 1 GCGUUCCUCAUCCUCCUUCGAUAUGGCCGAGUCCGGAUUGUACUUUGAUUCGGCCUUAUCCUUUGGAUGUCGUAUGACCUCAAUGACCUAAGAUCAAUUGAUAUAU----UA .(((.(..(((((......((((.(((((((((.((......))..))))))))).))))....)))))..).)))...(((((((.......)).))))).....----.. ( -28.50) >DroWil_CAF1 34216 106 - 1 ACGUUCUUCAUCCUCUUUCGAUAUAGCCGAAUCGGGAUUAUAUUUCGAUUCAGCUUUAUCUUUAGGAUGUCUUAUGACCUCAAUGACCUU--A--AAAAAAU--AGUUUUUA .(((....((((((.....((((.(((.((((((..((...))..)))))).))).))))...))))))....)))..............--.--.......--........ ( -25.80) >DroMoj_CAF1 22854 105 - 1 GCGUUCCUCAUCCUCUUUGGAUAUGGCCGAGUCUGGAUUAUACUUGGAUUCGGCUUUAUCUUUUGGAUGUCGUAUGACCUCAAUGACCUGAAA---AUUAAUAUAU----AC .(((.(..(((((.....(((((.(((((((((..(.......)..))))))))).)))))...)))))..).))).................---..........----.. ( -30.70) >DroAna_CAF1 19813 99 - 1 GCGUUCCUCAUCCUCUUUCGAUAUGGCUGAAUCAGGAUUAUACUUCGAUUCAGCUUUAUCCUUCGGAUGUCGUAUGACCUCAAUUACCUA-------GAGAU--UG----GU .(((.(..(((((......((((.((((((((((((......))).))))))))).))))....)))))..).)))...((((((.....-------..)))--))----). ( -26.50) >DroPer_CAF1 31407 99 - 1 GCGUUCUUCAUCCUCCUUCGAUAUGGCCGAAUCGGGAUUGUAUUUCGACUCGGCUUUAUCCUUGGGAUGUCGUAUGACCUCAAUGACCUG--C-GAA----A--GG----CA ..((..(((....((((..((((.((((((.(((..((...))..))).)))))).))))...)))).(((((.((....)))))))...--.-)))----.--.)----). ( -33.20) >consensus GCGUUCCUCAUCCUCUUUCGAUAUGGCCGAAUCGGGAUUAUACUUCGAUUCGGCUUUAUCCUUGGGAUGUCGUAUGACCUCAAUGACCUG__A__AAUAGAU__AG____CA .(((....(((((......((((.((((((((((((.......)))))))))))).))))....)))))....))).................................... (-24.83 = -24.55 + -0.28)

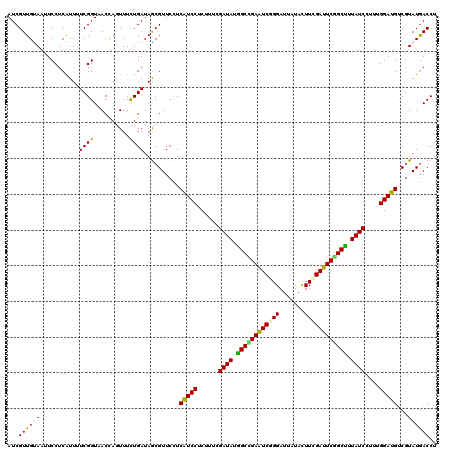
| Location | 15,026,682 – 15,026,802 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -26.72 |
| Energy contribution | -25.83 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15026682 120 - 22407834 AUCGUUGCAAUUCCUCGUUUUCGGUAACAAGUUUCUGAUAGCGUUCUUCGUCCUCUUUCGAUAUGGCUGAAUCGGGAUUGUAUUUCGAUUCGGCCUUAUCCUUGGGAUGACGUAUAACCU .....(((.......((((.((((.((....)).)))).))))....(((((((.....((((.((((((((((..((...))..)))))))))).))))...))))))).)))...... ( -38.20) >DroVir_CAF1 18821 120 - 1 AUCGUUGUAAUUCCUCAUUUUCGGUUACCAGUUUUUGAUAGCGUUCCUCGUCCUCCUUCGAUAUGGCCGAGUCUGGAUUGUACUUGGAUUCGGCUUUAUCUUUUGGAUGUCGUAUGACCU ......................(((((...(((......)))...(..(((((......((((.(((((((((..(.......)..))))))))).))))....)))))..)..))))). ( -31.10) >DroGri_CAF1 18911 120 - 1 AUCGUUGUAGUUCCUCGUUUUCGGUUACUAGUUUUUGAUAGCGUUCCUCAUCCUCCUUCGAUAUGGCCGAGUCCGGAUUGUACUUUGAUUCGGCCUUAUCCUUUGGAUGUCGUAUGACCU ...((..((......((((.((((..(....)..)))).))))..(..(((((......((((.(((((((((.((......))..))))))))).))))....)))))..)))..)).. ( -31.70) >DroWil_CAF1 34242 120 - 1 AUCGUUGUAGUUCCUCAUUCUCGGUAACCAAUUUCUGAUAACGUUCUUCAUCCUCUUUCGAUAUAGCCGAAUCGGGAUUAUAUUUCGAUUCAGCUUUAUCUUUAGGAUGUCUUAUGACCU ..((((((.(((((........)).)))((.....)))))))).....((((((.....((((.(((.((((((..((...))..)))))).))).))))...))))))........... ( -28.50) >DroMoj_CAF1 22879 120 - 1 AUCGUUGUAGUUCCUCGUUUUCGGUUACUAGUUUUUGAUAGCGUUCCUCAUCCUCUUUGGAUAUGGCCGAGUCUGGAUUAUACUUGGAUUCGGCUUUAUCUUUUGGAUGUCGUAUGACCU ...((..((......((((.((((..(....)..)))).))))..(..(((((.....(((((.(((((((((..(.......)..))))))))).)))))...)))))..)))..)).. ( -35.50) >DroAna_CAF1 19832 120 - 1 AUCGUUGCAAUUCCUCAUUUUCGGUAACCAAUUUCUGAUAGCGUUCCUCAUCCUCUUUCGAUAUGGCUGAAUCAGGAUUAUACUUCGAUUCAGCUUUAUCCUUCGGAUGUCGUAUGACCU ...(((((...............))))).............(((.(..(((((......((((.((((((((((((......))).))))))))).))))....)))))..).))).... ( -26.96) >consensus AUCGUUGUAAUUCCUCAUUUUCGGUAACCAGUUUCUGAUAGCGUUCCUCAUCCUCUUUCGAUAUGGCCGAAUCGGGAUUAUACUUCGAUUCGGCUUUAUCCUUUGGAUGUCGUAUGACCU ...((((((...........((((..........))))..........(((((......((((.((((((((((((.......)))))))))))).))))....)))))...)))))).. (-26.72 = -25.83 + -0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:10 2006