
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,996,588 – 14,996,762 |
| Length | 174 |
| Max. P | 0.901295 |

| Location | 14,996,588 – 14,996,684 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
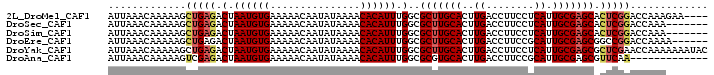
>2L_DroMel_CAF1 14996588 96 - 22407834 AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCUCAUUGCGAGCACUCGGACCAAAGAA---- .............(((((.(.((((((...............)))))).)..(((((((..(((......))).))))))).))))).........---- ( -21.76) >DroSec_CAF1 84368 93 - 1 AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCUCAUUGCGAGCACUCGGACCAAA------- .............(((((.(.((((((...............)))))).)..(((((((..(((......))).))))))).)))))......------- ( -21.76) >DroSim_CAF1 83407 93 - 1 AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCUCAUUGCGAGCACUCGGACCAAA------- .............(((((.(.((((((...............)))))).)..(((((((..(((......))).))))))).)))))......------- ( -21.76) >DroEre_CAF1 96198 94 - 1 AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCGCAUUGCGAGCGGCCGGACCAAAA------ .......................((((...............))))(((((((((((((..((........)).))))))).))))))......------ ( -18.46) >DroYak_CAF1 95230 100 - 1 AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCUCAUUGCGAGCGCUCGAACCAAAAAAAUAC ...................(.((((((...............)))))).)(((((((((..(((......))).)))))))))................. ( -20.46) >DroAna_CAF1 91328 87 - 1 AUUAAACAAAAAGUCGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCGUGCACUUGACCUUCCGCAUUGCGAGCGUUCAA------------- ............((((((.(.((((((...............)))))).)((....)).))))))....(((.......))).....------------- ( -13.56) >consensus AUUAAACAAAAAGCUGAGACUAAUGUGAAAAACAAUAUAAAACACAUUUGGCGCUUGCACUUGACCUUCCUCAUUGCGAGCACUCGGACCAAA_______ .............(((((.(.((((((...............)))))).)..(((((((..((........)).))))))).)))))............. (-16.64 = -17.14 + 0.50)
| Location | 14,996,651 – 14,996,762 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
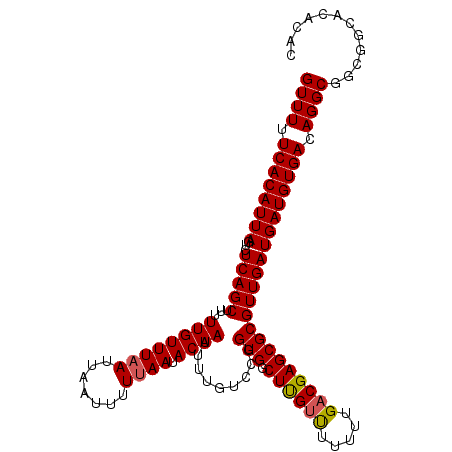
>2L_DroMel_CAF1 14996651 111 + 22407834 GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUUAAUACAAUUUUGGCCGGUGGCUUGUUUUGUUGACGAGCGCGUUGAUGAUGUGACAGGCGGCGGCACACAC ((((.((((((((......(((..((((((((.......)))).))))....)))(..(((((((((.....))))))).))..).)))))))).))))............ ( -26.70) >DroSec_CAF1 84428 111 + 1 GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUUAAUACAAUUUUGUGCGGUGGCUUGUUUUUUUGACGAGCGCGUUGAUGAUGUGACAGGCGGCGGCACACAC ((.......((((((...(((.....)))..)))))).......)).....(((((.((.(((((((...(..(((....)))..)......))))))).)).)))))... ( -27.34) >DroSim_CAF1 83467 111 + 1 GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUUAAUACAAUUUUGUGCGGUGGCUUGUUUUUUUGACGAGCGCGUUGAUGAUGUGACAGGCGGCGGCACACAC ((.......((((((...(((.....)))..)))))).......)).....(((((.((.(((((((...(..(((....)))..)......))))))).)).)))))... ( -27.34) >DroEre_CAF1 96259 108 + 1 GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUGAAUACAAUUUGGUCCGGUGGCUCGUUUUUUUGACGAGCGCGUUGAUGAUGUGACAGGC---GGCACACAC ((((.((((((((...(((((....(((((((......)))))))............(((.((((((.....)))))))))))))))))))))).))))---......... ( -29.30) >DroYak_CAF1 95297 107 + 1 GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUUAAUACAAUUUUGCUCGGUGGCUUGCCU-UUUGACGAGCGCGUUGAUGAUGUGACAGGC---GGCACACAC ((((.((((((((...(((((...((((((((.......)))).))))....(((((..((....)).-.....)))))..))))))))))))).))))---......... ( -26.60) >consensus GUUUUUCACAUUAGUCUCAGCUUUUUGUUUAAUUAAUUUUUAAUACAAUUUUGUCCGGUGGCUUGUUUUUUUGACGAGCGCGUUGAUGAUGUGACAGGCGGCGGCACACAC ((((.((((((((...(((((...((((((((.......)))).)))).........(((.((((((.....)))))))))))))))))))))).))))............ (-24.46 = -24.54 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:02 2006