
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,973,944 – 14,974,070 |
| Length | 126 |
| Max. P | 0.940646 |

| Location | 14,973,944 – 14,974,045 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -21.59 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14973944 101 + 22407834 AAUAACCUUCGCAGCAAUUACAGGCAGAAAACUGACACAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCG ..........((((((....(((........))).......................-....------((((((((.((.....)))))))))).))))))....-... ( -22.40) >DroSec_CAF1 69276 101 + 1 AAUAACCUUCGCAGCAAUUACAGGCAGAAAACUGACAAAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCG ..........((((((....(((........))).......................-....------((((((((.((.....)))))))))).))))))....-... ( -22.40) >DroSim_CAF1 68340 101 + 1 AAUAACCUUCGCAGCAAUUACAGGCAGAAAACUGACAAAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCG ..........((((((....(((........))).......................-....------((((((((.((.....)))))))))).))))))....-... ( -22.40) >DroEre_CAF1 80996 78 + 1 ACUAACCUUCGCUGCAAUUACAGGC-----------------------AAAAUUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCGGCUGCAGUU-GCG .............(((((((((((.-----------------------(((......-....------...)))))))).((((.(((.....))).))))))))-)). ( -19.42) >DroYak_CAF1 79290 101 + 1 AACAACCUUCACAGCAAUUACAGGCAGAAAACUGACAGAAAAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCG .............((((((.(((........)))............(((........-...(------(((.....))))(((((((....))))))))))))))-)). ( -21.90) >DroAna_CAF1 77602 96 + 1 CAUAACCUUCACAGCAAUUACAAGCAAA-AA------------CGAGCAAAACUCAAAAUAAAAAAUAUCAUUUCCUGCUGCAGCCGGAAACGUUUGUUGCAGCUAGCG .......................((...-..------------.(((.....)))......................((((((((((....))...))))))))..)). ( -21.00) >consensus AAUAACCUUCGCAGCAAUUACAGGCAGAAAACUGACA_AACAAUAAGCAAAAAUCAA_AUAA______ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU_GCG .........(((.((........))...................................................(((.(((((((....))))))).)))....))) (-13.20 = -13.32 + 0.11)

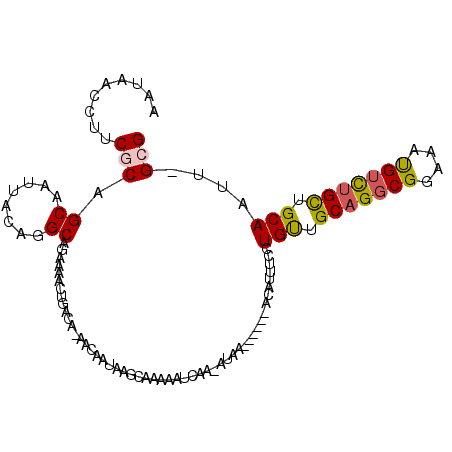

| Location | 14,973,944 – 14,974,045 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14973944 101 - 22407834 CGC-AAUUGCAGCAGACAUUUCCGCCUGCAACAGGAAAUGU------UUAU-UUGAUUUUUGCUUAUUGUUGUGUCAGUUUUCUGCCUGUAAUUGCUGCGAAGGUUAUU .((-(((((((((((((((((((..........))))))).------....-(((((....((.....))...)))))...)))).)))))))))).((....)).... ( -30.70) >DroSec_CAF1 69276 101 - 1 CGC-AAUUGCAGCAGACAUUUCCGCCUGCAACAGGAAAUGU------UUAU-UUGAUUUUUGCUUAUUGUUUUGUCAGUUUUCUGCCUGUAAUUGCUGCGAAGGUUAUU .((-(((((((((((((((((((..........))))))).------....-(((((....((.....))...)))))...)))).)))))))))).((....)).... ( -30.70) >DroSim_CAF1 68340 101 - 1 CGC-AAUUGCAGCAGACAUUUCCGCCUGCAACAGGAAAUGU------UUAU-UUGAUUUUUGCUUAUUGUUUUGUCAGUUUUCUGCCUGUAAUUGCUGCGAAGGUUAUU .((-(((((((((((((((((((..........))))))).------....-(((((....((.....))...)))))...)))).)))))))))).((....)).... ( -30.70) >DroEre_CAF1 80996 78 - 1 CGC-AACUGCAGCCGACAUUUCCGCCUGCAACAGGAAAUGU------UUAU-UUGAAUUUU-----------------------GCCUGUAAUUGCAGCGAAGGUUAGU .((-((.(((((.((.......)).)))))(((((....((------((..-..))))...-----------------------.)))))..))))(((....)))... ( -21.40) >DroYak_CAF1 79290 101 - 1 CGC-AAUUGCAGCAGACAUUUCCGCCUGCAACAGGAAAUGU------UUAU-UUGAUUUUUGCUUAUUUUUCUGUCAGUUUUCUGCCUGUAAUUGCUGUGAAGGUUGUU .((-(((((((((((((((((((..........))))))).------....-(((((................)))))...)))).))))))))))............. ( -27.49) >DroAna_CAF1 77602 96 - 1 CGCUAGCUGCAACAAACGUUUCCGGCUGCAGCAGGAAAUGAUAUUUUUUAUUUUGAGUUUUGCUCG------------UU-UUUGCUUGUAAUUGCUGUGAAGGUUAUG (((.(((((((((((((((((((.((....)).)))))))..............(((.....))).------------..-)))).)))))...))))))......... ( -23.30) >consensus CGC_AAUUGCAGCAGACAUUUCCGCCUGCAACAGGAAAUGU______UUAU_UUGAUUUUUGCUUAUUGUU_UGUCAGUUUUCUGCCUGUAAUUGCUGCGAAGGUUAUU (((.(((((((((((((((((((..........))))))).........................((((......))))..)))).))))))))...)))......... (-17.98 = -18.17 + 0.19)

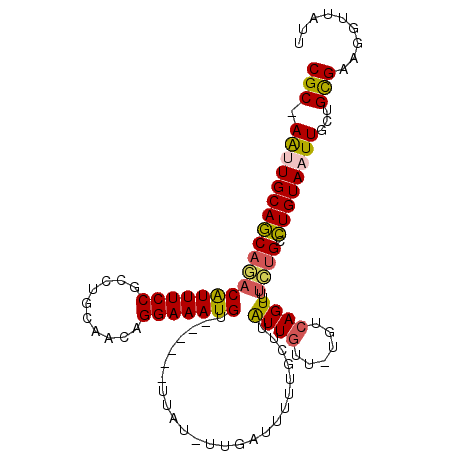
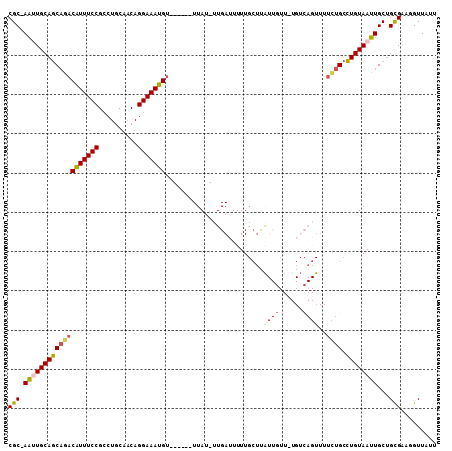
| Location | 14,973,977 – 14,974,070 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14973977 93 + 22407834 GACACAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCGCCCAAGCCCGCG-GCACACUGCGUAU .............(((........-....------((((((((.((.....)))))))))).((((((....-((......))..)))-)))...))).... ( -21.80) >DroSec_CAF1 69309 93 + 1 GACAAAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCGCCCAUGCCCGCG-GCACACUGCGUAU .............(((........-...(------(((.....))))(((((((....))))))))))...(-((((...((((...)-)))....))))). ( -22.40) >DroSim_CAF1 68373 93 + 1 GACAAAACAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCGCCCAAGCCCGCG-GCACACUGCGUAU .............(((........-....------((((((((.((.....)))))))))).((((((....-((......))..)))-)))...))).... ( -21.80) >DroEre_CAF1 81021 78 + 1 ---------------AAAAUUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCGGCUGCAGUU-GCGCCCAAGCCCGCG-CAACACUGCGUAU ---------------.........-....------((((((((.((.....)))))))))).....((((((-((((........)))-))..))))).... ( -23.20) >DroYak_CAF1 79323 93 + 1 GACAGAAAAAUAAGCAAAAAUCAA-AUAA------ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU-GCGCCCAGGCCCGCG-GCACACUGCGUAU .............(((........-...(------(((.....))))(((((((....))))))))))...(-((((....(((...)-)).....))))). ( -21.90) >DroAna_CAF1 77632 92 + 1 ----------CGAGCAAAACUCAAAAUAAAAAAUAUCAUUUCCUGCUGCAGCCGGAAACGUUUGUUGCAGCUAGCGCCCAACCUGGGGACCACGUUGCGUAU ----------.(((.....)))......................((((((((((....))...))))))))..((((..(((.(((...))).))))))).. ( -25.60) >consensus GACA_AACAAUAAGCAAAAAUCAA_AUAA______ACAUUUCCUGUUGCAGGCGGAAAUGUCUGCUGCAAUU_GCGCCCAAGCCCGCG_GCACACUGCGUAU .............(((...........................(((.(((((((....))))))).)))....(((........)))........))).... (-14.10 = -14.55 + 0.45)
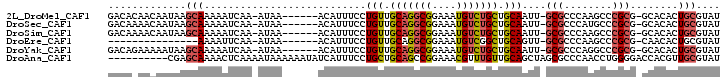

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:58 2006